| 1 |
|
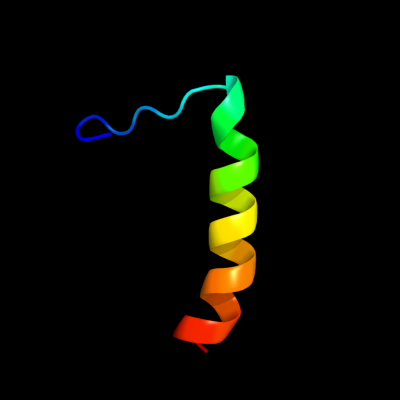
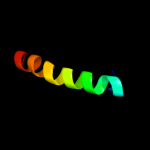
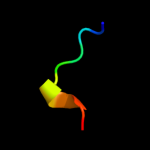
PDB 2bo9 chain B domain 2
Region: 12 - 37
Aligned: 26
Modelled: 26
Confidence: 27.6%
Identity: 19%
Fold: Cystatin-like
Superfamily: Cystatin/monellin
Family: Latexin-like
Phyre2
| 2 |
|
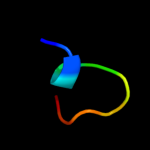
PDB 1pcf chain A
Region: 9 - 19
Aligned: 11
Modelled: 11
Confidence: 18.1%
Identity: 18%
Fold: ssDNA-binding transcriptional regulator domain
Superfamily: ssDNA-binding transcriptional regulator domain
Family: Transcriptional coactivator PC4 C-terminal domain
Phyre2
| 3 |
|
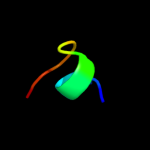
PDB 1cks chain A
Region: 2 - 14
Aligned: 13
Modelled: 13
Confidence: 14.0%
Identity: 23%
Fold: Cell cycle regulatory proteins
Superfamily: Cell cycle regulatory proteins
Family: Cell cycle regulatory proteins
Phyre2
| 4 |
|
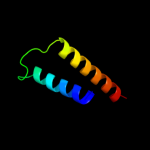
PDB 2bo9 chain B
Region: 12 - 40
Aligned: 29
Modelled: 29
Confidence: 13.2%
Identity: 17%
PDB header:hydrolase
Chain: B: PDB Molecule:human latexin;
PDBTitle: human carboxypeptidase a4 in complex with human latexin.
Phyre2
| 5 |
|
PDB 2lma chain A
Region: 9 - 20
Aligned: 12
Modelled: 12
Confidence: 12.5%
Identity: 25%
PDB header:immune system
Chain: A: PDB Molecule:thp5 peptide;
PDBTitle: solution structure of cd4+ t cell derived peptide thp5
Phyre2
| 6 |
|
PDB 3bzk chain A domain 2
Region: 3 - 12
Aligned: 10
Modelled: 10
Confidence: 11.5%
Identity: 30%
Fold: SAM domain-like
Superfamily: RuvA domain 2-like
Family: Tex HhH-containing domain-like
Phyre2
| 7 |
|
PDB 2k9p chain A
Region: 61 - 110
Aligned: 49
Modelled: 50
Confidence: 11.1%
Identity: 10%
PDB header:membrane protein
Chain: A: PDB Molecule:pheromone alpha factor receptor;
PDBTitle: structure of tm1_tm2 in lppg micelles
Phyre2
| 8 |
|
PDB 2ast chain C domain 1
Region: 3 - 14
Aligned: 12
Modelled: 12
Confidence: 9.5%
Identity: 8%
Fold: Cell cycle regulatory proteins
Superfamily: Cell cycle regulatory proteins
Family: Cell cycle regulatory proteins
Phyre2
| 9 |
|
PDB 1puc chain A
Region: 2 - 14
Aligned: 13
Modelled: 13
Confidence: 8.8%
Identity: 15%
Fold: Cell cycle regulatory proteins
Superfamily: Cell cycle regulatory proteins
Family: Cell cycle regulatory proteins
Phyre2
| 10 |
|
PDB 1s6l chain A domain 1
Region: 11 - 17
Aligned: 7
Modelled: 7
Confidence: 7.8%
Identity: 43%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: MerB N-terminal domain-like
Phyre2
| 11 |
|
PDB 3qao chain A
Region: 10 - 20
Aligned: 11
Modelled: 11
Confidence: 7.5%
Identity: 45%
PDB header:transcription regulator
Chain: A: PDB Molecule:merr-like transcriptional regulator;
PDBTitle: the crystal structure of the n-terminal domain of a merr-like2 transcriptional regulator from listeria monocytogenes egd-e
Phyre2
| 12 |
|
PDB 1mg7 chain A domain 2
Region: 136 - 149
Aligned: 14
Modelled: 14
Confidence: 6.7%
Identity: 21%
Fold: Ferredoxin-like
Superfamily: GHMP Kinase, C-terminal domain
Family: Early switch protein XOL-1
Phyre2
| 13 |
|
PDB 1qb3 chain B
Region: 2 - 14
Aligned: 13
Modelled: 13
Confidence: 6.5%
Identity: 23%
PDB header:cell cycle
Chain: B: PDB Molecule:cyclin-dependent kinases regulatory subunit;
PDBTitle: crystal structure of the cell cycle regulatory protein cks1
Phyre2
| 14 |
|
PDB 1y2t chain A
Region: 2 - 12
Aligned: 11
Modelled: 11
Confidence: 6.1%
Identity: 9%
Fold: Cytolysin/lectin
Superfamily: Cytolysin/lectin
Family: Fungal fruit body lectin
Phyre2
| 15 |
|
PDB 2oce chain A
Region: 3 - 12
Aligned: 10
Modelled: 10
Confidence: 5.9%
Identity: 30%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein pa5201;
PDBTitle: crystal structure of tex family protein pa5201 from2 pseudomonas aeruginosa
Phyre2
| 16 |
|
PDB 1x99 chain A
Region: 7 - 16
Aligned: 10
Modelled: 10
Confidence: 5.7%
Identity: 40%
Fold: Cytolysin/lectin
Superfamily: Cytolysin/lectin
Family: Fungal fruit body lectin
Phyre2
| 17 |
|
PDB 1qb3 chain A
Region: 2 - 14
Aligned: 13
Modelled: 13
Confidence: 5.7%
Identity: 23%
Fold: Cell cycle regulatory proteins
Superfamily: Cell cycle regulatory proteins
Family: Cell cycle regulatory proteins
Phyre2
| 18 |
|
PDB 3d6z chain A
Region: 10 - 19
Aligned: 10
Modelled: 10
Confidence: 5.4%
Identity: 20%
PDB header:transcription regulator/dna
Chain: A: PDB Molecule:multidrug-efflux transporter 1 regulator;
PDBTitle: crystal structure of r275e mutant of bmrr bound to dna and rhodamine
Phyre2