| 1 |
|
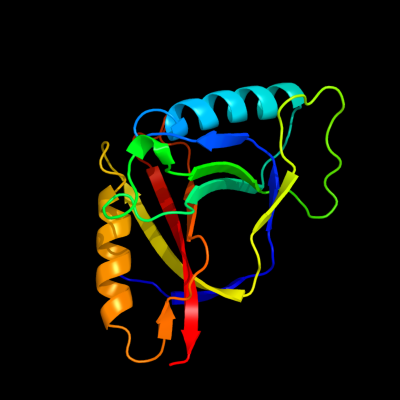
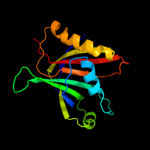
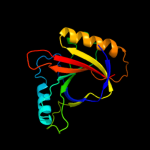
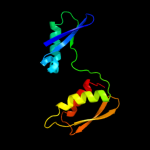
PDB 1jyh chain A
Region: 1 - 155
Aligned: 155
Modelled: 155
Confidence: 100.0%
Identity: 100%
Fold: Probable bacterial effector-binding domain
Superfamily: Probable bacterial effector-binding domain
Family: Gyrase inhibitory protein GyrI (SbmC, YeeB)
Phyre2
| 2 |
|
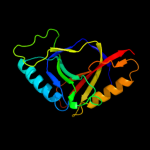
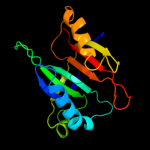
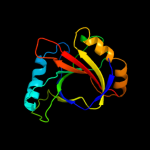
PDB 3lur chain B
Region: 1 - 153
Aligned: 149
Modelled: 152
Confidence: 100.0%
Identity: 13%
PDB header:transcription activator
Chain: B: PDB Molecule:putative bacterial transcription regulation protein;
PDBTitle: crystal structure of putative bacterial transcription regulation2 protein (np_372959.1) from staphylococcus aureus mu50 at 1.81 a3 resolution
Phyre2
| 3 |
|
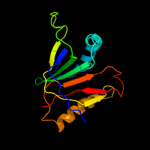
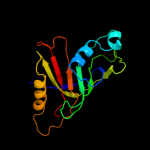
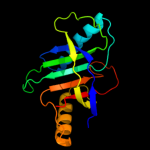
PDB 1d5y chain A domain 3
Region: 1 - 155
Aligned: 154
Modelled: 155
Confidence: 100.0%
Identity: 8%
Fold: Probable bacterial effector-binding domain
Superfamily: Probable bacterial effector-binding domain
Family: Rob transcription factor, C-terminal domain
Phyre2
| 4 |
|
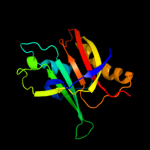
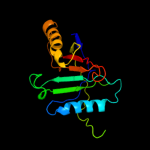
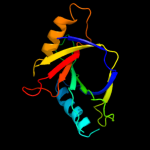
PDB 1d5y chain D
Region: 1 - 155
Aligned: 154
Modelled: 155
Confidence: 100.0%
Identity: 8%
PDB header:transcription/dna
Chain: D: PDB Molecule:rob transcription factor;
PDBTitle: crystal structure of the e. coli rob transcription factor2 in complex with dna
Phyre2
| 5 |
|
PDB 3gk6 chain A
Region: 1 - 153
Aligned: 140
Modelled: 150
Confidence: 100.0%
Identity: 16%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:integron cassette protein vch_cass2;
PDBTitle: crystal structure from the mobile metagenome of vibrio cholerae.2 integron cassette protein vch_cass2.
Phyre2
| 6 |
|
PDB 2kcu chain A
Region: 1 - 154
Aligned: 146
Modelled: 154
Confidence: 100.0%
Identity: 12%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein ctr107;
PDBTitle: nmr solution structure of an uncharacterized protein from2 chlorobium tepidum. northeast structural genomics target3 ctr107
Phyre2
| 7 |
|
PDB 3b49 chain A
Region: 2 - 152
Aligned: 150
Modelled: 151
Confidence: 99.9%
Identity: 10%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lin2189 protein;
PDBTitle: crystal structure of an uncharacterized conserved protein from2 listeria innocua
Phyre2
| 8 |
|
PDB 1r8e chain A domain 2
Region: 2 - 152
Aligned: 149
Modelled: 151
Confidence: 99.8%
Identity: 11%
Fold: Probable bacterial effector-binding domain
Superfamily: Probable bacterial effector-binding domain
Family: Multidrug-binding domain of transcription activator BmrR
Phyre2
| 9 |
|
PDB 3d6z chain A
Region: 2 - 153
Aligned: 150
Modelled: 152
Confidence: 99.7%
Identity: 11%
PDB header:transcription regulator/dna
Chain: A: PDB Molecule:multidrug-efflux transporter 1 regulator;
PDBTitle: crystal structure of r275e mutant of bmrr bound to dna and rhodamine
Phyre2
| 10 |
|
PDB 3r8k chain B
Region: 2 - 153
Aligned: 150
Modelled: 152
Confidence: 97.8%
Identity: 15%
PDB header:apoptosis
Chain: B: PDB Molecule:heme-binding protein 2;
PDBTitle: crystal structure of human soul protein (hexagonal form)
Phyre2
| 11 |
|
PDB 2gov chain A domain 1
Region: 2 - 150
Aligned: 147
Modelled: 149
Confidence: 97.7%
Identity: 14%
Fold: Probable bacterial effector-binding domain
Superfamily: Probable bacterial effector-binding domain
Family: SOUL heme-binding protein
Phyre2
| 12 |
|
PDB 2z51 chain A
Region: 2 - 120
Aligned: 116
Modelled: 119
Confidence: 10.8%
Identity: 11%
PDB header:metal transport
Chain: A: PDB Molecule:nifu-like protein 2, chloroplast;
PDBTitle: crystal structure of arabidopsis cnfu involved in iron-2 sulfur cluster biosynthesis
Phyre2