| 1 |
|
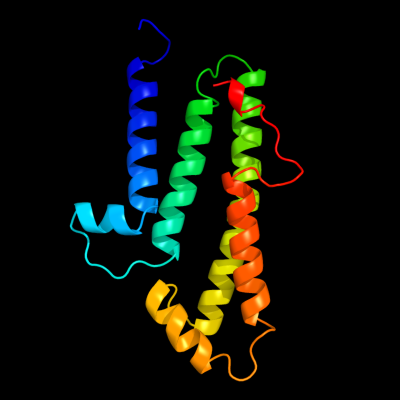
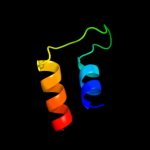
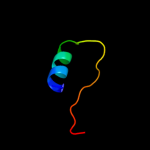
PDB 3rko chain L
Region: 58 - 203
Aligned: 136
Modelled: 146
Confidence: 95.6%
Identity: 15%
PDB header:oxidoreductase
Chain: L: PDB Molecule:nadh-quinone oxidoreductase subunit l;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 2 |
|
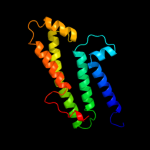
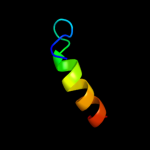
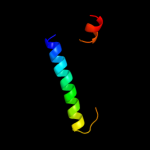
PDB 2jpm chain A
Region: 38 - 62
Aligned: 25
Modelled: 25
Confidence: 37.8%
Identity: 16%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:bacteriocin lactococcin-g subunit beta;
PDBTitle: lactococcin g-b in tfe
Phyre2
| 3 |
|
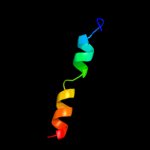
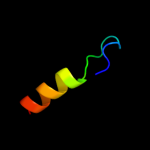
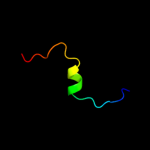
PDB 1q2i chain A
Region: 37 - 52
Aligned: 16
Modelled: 16
Confidence: 34.2%
Identity: 44%
PDB header:antitumor protein
Chain: A: PDB Molecule:pnc27;
PDBTitle: nmr solution structure of a peptide from the mdm-2 binding2 domain of the p53 protein that is selectively cytotoxic to3 cancer cells
Phyre2
| 4 |
|
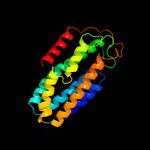
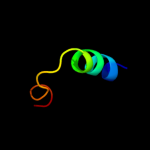
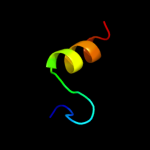
PDB 1lbq chain A
Region: 32 - 53
Aligned: 22
Modelled: 22
Confidence: 32.8%
Identity: 27%
Fold: Chelatase-like
Superfamily: Chelatase
Family: Ferrochelatase
Phyre2
| 5 |
|
PDB 3rko chain M
Region: 71 - 273
Aligned: 194
Modelled: 203
Confidence: 30.1%
Identity: 11%
PDB header:oxidoreductase
Chain: M: PDB Molecule:nadh-quinone oxidoreductase subunit m;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 6 |
|
PDB 1xn8 chain A
Region: 195 - 229
Aligned: 35
Modelled: 35
Confidence: 25.7%
Identity: 17%
Fold: Hypothetical protein YqbG
Superfamily: Hypothetical protein YqbG
Family: Hypothetical protein YqbG
Phyre2
| 7 |
|
PDB 2hrc chain A domain 1
Region: 32 - 53
Aligned: 22
Modelled: 22
Confidence: 15.7%
Identity: 27%
Fold: Chelatase-like
Superfamily: Chelatase
Family: Ferrochelatase
Phyre2
| 8 |
|
PDB 2hk6 chain A domain 1
Region: 32 - 53
Aligned: 22
Modelled: 22
Confidence: 11.1%
Identity: 23%
Fold: Chelatase-like
Superfamily: Chelatase
Family: Ferrochelatase
Phyre2
| 9 |
|
PDB 2dbh chain A
Region: 24 - 45
Aligned: 22
Modelled: 22
Confidence: 10.2%
Identity: 23%
PDB header:signaling protein
Chain: A: PDB Molecule:tumor necrosis factor receptor superfamily
PDBTitle: solution structure of the carboxyl-terminal card-like2 domain in human tnfr-related death receptor-6
Phyre2
| 10 |
|
PDB 2vxs chain B
Region: 191 - 201
Aligned: 11
Modelled: 10
Confidence: 8.3%
Identity: 27%
PDB header:cytokine
Chain: B: PDB Molecule:interleukin-17a;
PDBTitle: structure of il-17a in complex with a potent, fully human2 neutralising antibody
Phyre2
| 11 |
|
PDB 1ufr chain A
Region: 195 - 214
Aligned: 20
Modelled: 20
Confidence: 7.9%
Identity: 15%
Fold: PRTase-like
Superfamily: PRTase-like
Family: Phosphoribosyltransferases (PRTases)
Phyre2
| 12 |
|
PDB 3dpg chain A
Region: 30 - 46
Aligned: 17
Modelled: 17
Confidence: 7.3%
Identity: 29%
PDB header:hydrolase/dna
Chain: A: PDB Molecule:sgrair restriction enzyme;
PDBTitle: sgrai with noncognate dna bound
Phyre2
| 13 |
|
PDB 1jpy chain A
Region: 191 - 201
Aligned: 11
Modelled: 11
Confidence: 5.8%
Identity: 18%
Fold: Cystine-knot cytokines
Superfamily: Cystine-knot cytokines
Family: Interleukin 17F, IL-17F
Phyre2
| 14 |
|
PDB 3eto chain B
Region: 180 - 205
Aligned: 26
Modelled: 26
Confidence: 5.7%
Identity: 19%
PDB header:signaling protein
Chain: B: PDB Molecule:neurogenic locus notch homolog protein 1;
PDBTitle: 2 angstrom xray structure of the notch1 negative regulatory region2 (nrr)
Phyre2
| 15 |
|
PDB 1eys chain M
Region: 9 - 46
Aligned: 38
Modelled: 38
Confidence: 5.6%
Identity: 13%
Fold: Bacterial photosystem II reaction centre, L and M subunits
Superfamily: Bacterial photosystem II reaction centre, L and M subunits
Family: Bacterial photosystem II reaction centre, L and M subunits
Phyre2
| 16 |
|
PDB 2jz8 chain A
Region: 193 - 208
Aligned: 16
Modelled: 16
Confidence: 5.6%
Identity: 25%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein bh09830;
PDBTitle: solution nmr structure of bh09830 from bartonella henselae2 modeled with one zn+2 bound. northeast structural genomics3 consortium target bnr55
Phyre2
| 17 |
|
PDB 2j8c chain M domain 1
Region: 9 - 46
Aligned: 38
Modelled: 38
Confidence: 5.6%
Identity: 13%
Fold: Bacterial photosystem II reaction centre, L and M subunits
Superfamily: Bacterial photosystem II reaction centre, L and M subunits
Family: Bacterial photosystem II reaction centre, L and M subunits
Phyre2
| 18 |
|
PDB 2k8f chain B
Region: 35 - 57
Aligned: 23
Modelled: 23
Confidence: 5.5%
Identity: 35%
PDB header:transferase/transcription
Chain: B: PDB Molecule:cellular tumor antigen p53;
PDBTitle: structural basis for the regulation of p53 function by p300
Phyre2
| 19 |
|
PDB 3llb chain A
Region: 196 - 215
Aligned: 20
Modelled: 20
Confidence: 5.3%
Identity: 15%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: the crystal structure of the protein pa3983 with unknown2 function from pseudomonas aeruginosa pao1
Phyre2