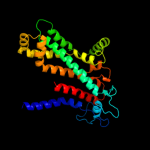
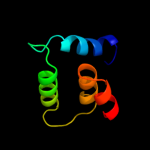
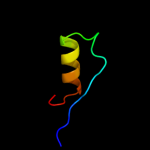
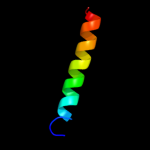
1 d3dhwa1
99.9
23
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like2 d2onkc1
99.8
12
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like3 c2onkC_
99.8
12
PDB header: membrane proteinChain: C: PDB Molecule: molybdate/tungstate abc transporter, permeasePDBTitle: abc transporter modbc in complex with its binding protein2 moda
4 c3d31D_
99.7
11
PDB header: transport proteinChain: D: PDB Molecule: sulfate/molybdate abc transporter, permeasePDBTitle: modbc from methanosarcina acetivorans
5 d3d31c1
99.7
11
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like6 d2r6gf2
99.6
12
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like7 c3fh6F_
99.6
15
PDB header: transport proteinChain: F: PDB Molecule: maltose transport system permease protein malf;PDBTitle: crystal structure of the resting state maltose transporter from e.2 coli
8 c2r6gF_
99.5
12
PDB header: hydrolase/transport proteinChain: F: PDB Molecule: maltose transport system permease protein malf;PDBTitle: the crystal structure of the e. coli maltose transporter
9 d2r6gg1
99.1
13
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like10 c1ciiA_
26.3
16
PDB header: transmembrane proteinChain: A: PDB Molecule: colicin ia;PDBTitle: colicin ia
11 c3ednB_
26.2
21
PDB header: biosynthetic proteinChain: B: PDB Molecule: phenazine biosynthesis protein, phzf family;PDBTitle: crystal structure of the bacillus anthracis phenazine2 biosynthesis protein, phzf family
12 d1qy9a1
26.2
17
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like13 c2ka1A_
24.4
12
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles
14 c2ka2B_
24.4
12
PDB header: membrane proteinChain: B: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles with his173-ser172 intermonomer3 hydrogen bond restraints
15 c2ka2A_
24.4
12
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles with his173-ser172 intermonomer3 hydrogen bond restraints
16 c2ka1B_
24.4
12
PDB header: membrane proteinChain: B: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles
17 d1xuba1
23.5
21
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like18 c1u0kA_
21.3
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: gene product pa4716;PDBTitle: the structure of a predicted epimerase pa4716 from pseudomonas2 aeruginosa
19 d1u0ka1
20.1
10
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like20 d1s7ja_
19.6
17
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like21 c2j5dA_
not modelled
19.5
12
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: nmr structure of bnip3 transmembrane domain in lipid2 bicelles
22 c1u1wA_
not modelled
19.4
21
PDB header: isomerase, lyaseChain: A: PDB Molecule: phenazine biosynthesis protein phzf;PDBTitle: structure and function of phenazine-biosynthesis protein phzf from2 pseudomonas fluorescens 2-79
23 c1wz4A_
not modelled
19.4
38
PDB header: gene regulationChain: A: PDB Molecule: major surface antigen;PDBTitle: solution conformation of adr subtype hbv pre-s2 epitope
24 c1qy9B_
not modelled
17.1
17
PDB header: unknown functionChain: B: PDB Molecule: hypothetical protein ydde;PDBTitle: crystal structure of e. coli se-met protein ydde
25 d2d6fc1
not modelled
16.9
15
Fold: GatB/YqeY motifSuperfamily: GatB/YqeY motifFamily: GatB/GatE C-terminal domain-like26 c2cw1A_
not modelled
16.1
32
PDB header: de novo proteinChain: A: PDB Molecule: sn4m;PDBTitle: solution structure of the de novo-designed lambda cro fold2 protein
27 c2iv1J_
not modelled
16.0
22
PDB header: lyaseChain: J: PDB Molecule: cyanate hydratase;PDBTitle: site directed mutagenesis of key residues involved in the2 catalytic mechanism of cyanase
28 c1y66D_
not modelled
13.6
21
PDB header: de novo proteinChain: D: PDB Molecule: engrailed homeodomain;PDBTitle: dioxane contributes to the altered conformation and2 oligomerization state of a designed engrailed homeodomain3 variant
29 c2hw2A_
not modelled
13.0
37
PDB header: transferaseChain: A: PDB Molecule: rifampin adp-ribosyl transferase;PDBTitle: crystal structure of rifampin adp-ribosyl transferase in2 complex with rifampin
30 d1a9xa1
not modelled
12.9
19
Fold: Carbamoyl phosphate synthetase, large subunit connection domainSuperfamily: Carbamoyl phosphate synthetase, large subunit connection domainFamily: Carbamoyl phosphate synthetase, large subunit connection domain31 c1ym5A_
not modelled
10.6
24
PDB header: oxidoreductaseChain: A: PDB Molecule: hypothetical 32.6 kda protein in dap2-slt2PDBTitle: crystal structure of yhi9, the yeast member of the2 phenazine biosynthesis phzf enzyme superfamily.
32 c2v9kA_
not modelled
9.6
32
PDB header: lyaseChain: A: PDB Molecule: uncharacterized protein flj32312;PDBTitle: crystal structure of human pus10, a novel pseudouridine2 synthase.
33 d1n0ua5
not modelled
8.8
23
Fold: Ferredoxin-likeSuperfamily: EF-G C-terminal domain-likeFamily: EF-G/eEF-2 domains III and V34 d2oara1
not modelled
8.3
13
Fold: Gated mechanosensitive channelSuperfamily: Gated mechanosensitive channelFamily: Gated mechanosensitive channel35 d1dwka1
not modelled
8.0
19
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Cyanase N-terminal domain36 d1g2ha_
not modelled
7.6
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like37 c2b9sB_
not modelled
6.7
38
PDB header: isomerase/dnaChain: B: PDB Molecule: dna topoisomerase i-like protein;PDBTitle: crystal structure of heterodimeric l. donovani2 topoisomerase i-vanadate-dna complex
38 c2oarA_
not modelled
6.6
13
PDB header: membrane proteinChain: A: PDB Molecule: large-conductance mechanosensitive channel;PDBTitle: mechanosensitive channel of large conductance (mscl)
39 c1umqA_
not modelled
6.4
19
PDB header: dna-binding proteinChain: A: PDB Molecule: photosynthetic apparatus regulatory protein;PDBTitle: solution structure and dna binding of the effector domain2 from the global regulator prra(rega) from r. sphaeroides:3 insights into dna binding specificity
40 d1umqa_
not modelled
6.4
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like41 d1ddfa_
not modelled
5.8
6
Fold: DEATH domainSuperfamily: DEATH domainFamily: DEATH domain, DD42 d2auwa1
not modelled
5.7
29
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: NE0471 C-terminal domain-like43 d1gt1a_
not modelled
5.5
18
Fold: LipocalinsSuperfamily: LipocalinsFamily: Retinol binding protein-like44 c2knaA_
not modelled
5.5
10
PDB header: apoptosisChain: A: PDB Molecule: baculoviral iap repeat-containing protein 4;PDBTitle: solution structure of uba domain of xiap