| 1 |
|
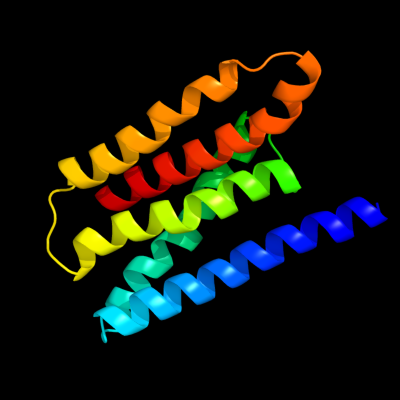
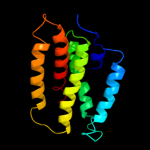
PDB 3k07 chain A
Region: 14 - 151
Aligned: 138
Modelled: 138
Confidence: 25.0%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:cation efflux system protein cusa;
PDBTitle: crystal structure of cusa
Phyre2
| 2 |
|
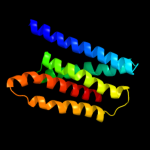
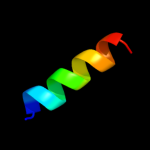
PDB 1iwg chain A domain 7
Region: 2 - 153
Aligned: 152
Modelled: 152
Confidence: 16.3%
Identity: 7%
Fold: Multidrug efflux transporter AcrB transmembrane domain
Superfamily: Multidrug efflux transporter AcrB transmembrane domain
Family: Multidrug efflux transporter AcrB transmembrane domain
Phyre2
| 3 |
|
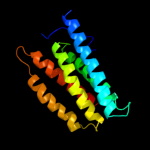
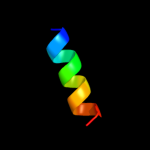
PDB 1oy8 chain A
Region: 13 - 153
Aligned: 141
Modelled: 141
Confidence: 9.7%
Identity: 5%
PDB header:membrane protein
Chain: A: PDB Molecule:acriflavine resistance protein b;
PDBTitle: structural basis of multiple drug binding capacity of the acrb2 multidrug efflux pump
Phyre2
| 4 |
|
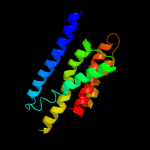
PDB 2e76 chain D
Region: 13 - 46
Aligned: 34
Modelled: 34
Confidence: 8.9%
Identity: 9%
PDB header:photosynthesis
Chain: D: PDB Molecule:cytochrome b6-f complex iron-sulfur subunit;
PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
Phyre2
| 5 |
|
PDB 1iwg chain A domain 8
Region: 5 - 150
Aligned: 146
Modelled: 146
Confidence: 7.3%
Identity: 12%
Fold: Multidrug efflux transporter AcrB transmembrane domain
Superfamily: Multidrug efflux transporter AcrB transmembrane domain
Family: Multidrug efflux transporter AcrB transmembrane domain
Phyre2
| 6 |
|
PDB 2caz chain A domain 1
Region: 11 - 23
Aligned: 13
Modelled: 13
Confidence: 7.1%
Identity: 15%
Fold: Long alpha-hairpin
Superfamily: Endosomal sorting complex assembly domain
Family: VPS23 C-terminal domain
Phyre2
| 7 |
|
PDB 2h3o chain A
Region: 115 - 130
Aligned: 16
Modelled: 16
Confidence: 7.1%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:merf;
PDBTitle: structure of merft, a membrane protein with two trans-2 membrane helices
Phyre2
| 8 |
|
PDB 2caz chain D
Region: 11 - 23
Aligned: 13
Modelled: 13
Confidence: 6.8%
Identity: 15%
PDB header:protein transport
Chain: D: PDB Molecule:suppressor protein stp22 of temperature-
PDBTitle: escrt-i core
Phyre2
| 9 |
|
PDB 2f6m chain A domain 1
Region: 10 - 23
Aligned: 14
Modelled: 14
Confidence: 6.7%
Identity: 14%
Fold: Long alpha-hairpin
Superfamily: Endosomal sorting complex assembly domain
Family: VPS23 C-terminal domain
Phyre2
| 10 |
|
PDB 1waz chain A
Region: 113 - 130
Aligned: 18
Modelled: 18
Confidence: 5.8%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:merf;
PDBTitle: nmr structure determination of the bacterial mercury2 transporter, merf, in micelles
Phyre2