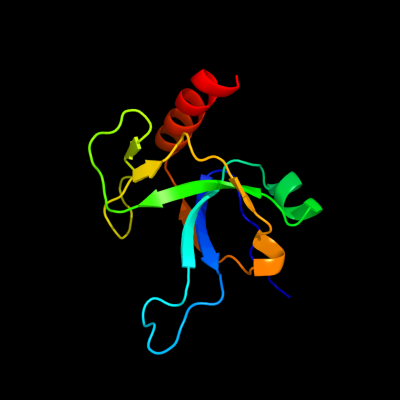
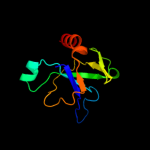
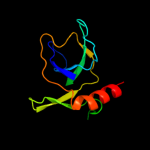
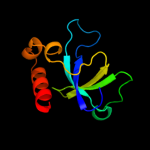
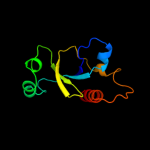
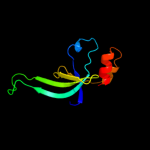
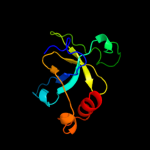
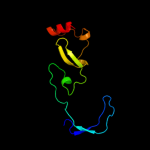
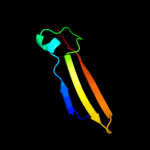
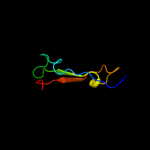
1 d1ub4a_
100.0
100
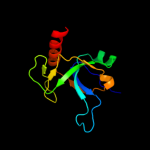
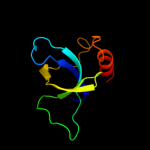
Fold: SH3-like barrelSuperfamily: Cell growth inhibitor/plasmid maintenance toxic componentFamily: Kid/PemK2 d1m1fa_
100.0
29
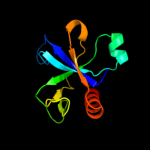
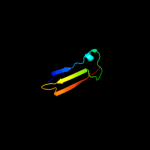
Fold: SH3-like barrelSuperfamily: Cell growth inhibitor/plasmid maintenance toxic componentFamily: Kid/PemK3 d1ne8a_
100.0
25
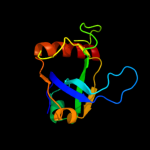
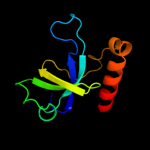
Fold: SH3-like barrelSuperfamily: Cell growth inhibitor/plasmid maintenance toxic componentFamily: Kid/PemK4 c3jrzA_
95.7
17
PDB header: toxinChain: A: PDB Molecule: ccdb;PDBTitle: ccdbvfi-formii-ph5.6
5 d3vuba_
94.9
15
Fold: SH3-like barrelSuperfamily: Cell growth inhibitor/plasmid maintenance toxic componentFamily: CcdB6 c3llrA_
70.5
14
PDB header: transferaseChain: A: PDB Molecule: dna (cytosine-5)-methyltransferase 3a;PDBTitle: crystal structure of the pwwp domain of human dna (cytosine-5-)-2 methyltransferase 3 alpha
7 d2daqa1
60.8
11
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: PWWP domain8 c2l89A_
50.5
19
PDB header: protein bindingChain: A: PDB Molecule: pwwp domain-containing protein 1;PDBTitle: solution structure of pdp1 pwwp domain reveals its unique binding2 sites for methylated h4k20 and dna
9 d1h3za_
48.8
10
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: PWWP domain10 d2nlua1
43.6
14
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: PWWP domain11 c3mxuA_
31.2
26
PDB header: oxidoreductaseChain: A: PDB Molecule: glycine cleavage system h protein;PDBTitle: crystal structure of glycine cleavage system protein h from bartonella2 henselae
12 d1cxqa_
27.9
12
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain13 d1n27a_
23.8
15
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: PWWP domain14 c2edgA_
23.5
21
PDB header: biosynthetic proteinChain: A: PDB Molecule: glycine cleavage system h protein;PDBTitle: solution structure of the gcv_h domain from mouse glycine
15 d1khca_
21.0
15
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: PWWP domain16 d2je6i1
20.9
27
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like17 c2h8kA_
20.5
29
PDB header: transferaseChain: A: PDB Molecule: sult1c3 splice variant d;PDBTitle: human sulfotranferase sult1c3 in complex with pap
18 c3a8jF_
20.2
29
PDB header: transferase/transport proteinChain: F: PDB Molecule: glycine cleavage system h protein;PDBTitle: crystal structure of et-ehred complex
19 d2ja9a1
19.1
33
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like20 d1onla_
18.4
24
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains21 c2x35A_
not modelled
16.7
13
PDB header: transcriptionChain: A: PDB Molecule: peregrin;PDBTitle: molecular basis of histone h3k36me3 recognition by the pwwp2 domain of brpf1.
22 c3pfsA_
not modelled
16.2
16
PDB header: protein bindingChain: A: PDB Molecule: bromodomain and phd finger-containing protein 3;PDBTitle: pwwp domain of human bromodomain and phd finger-containing protein 3
23 c2ja9A_
not modelled
15.7
33
PDB header: rna-binding proteinChain: A: PDB Molecule: exosome complex exonuclease rrp40;PDBTitle: structure of the n-terminal deletion of yeast exosome2 component rrp40
24 d2nn6g1
not modelled
14.4
55
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like25 c3iftA_
not modelled
14.2
21
PDB header: oxidoreductaseChain: A: PDB Molecule: glycine cleavage system h protein;PDBTitle: crystal structure of glycine cleavage system protein h from2 mycobacterium tuberculosis, using x-rays from the compact light3 source.
26 d2nn6i1
not modelled
13.8
31
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like27 d1aqua_
not modelled
13.4
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase28 d2ba0a1
not modelled
13.4
44
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like29 c2gfuA_
not modelled
13.0
18
PDB header: dna binding proteinChain: A: PDB Molecule: dna mismatch repair protein msh6;PDBTitle: nmr solution structure of the pwwp domain of mismatch2 repair protein hmsh6
30 d1vqoq1
not modelled
12.8
24
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e31 d1b12a_
not modelled
12.5
14
Fold: LexA/Signal peptidaseSuperfamily: LexA/Signal peptidaseFamily: Type 1 signal peptidase32 d1hpca_
not modelled
12.3
16
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains33 d2nn6h1
not modelled
11.9
36
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like34 d3bfxa1
not modelled
11.1
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase35 c2jnsA_
not modelled
10.8
14
PDB header: unknown functionChain: A: PDB Molecule: bromodomain-containing protein 4;PDBTitle: solution structure of the bromodomain-containing protein 42 et domain
36 d1xv1a_
not modelled
9.8
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase37 c2zvpX_
not modelled
9.6
16
PDB header: transferaseChain: X: PDB Molecule: tyrosine-ester sulfotransferase;PDBTitle: crystal structure of mouse cytosolic sulfotransferase msult1d1 complex2 with pap and p-nitrophenol
38 d1q44a_
not modelled
9.3
29
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase39 d1zaka2
not modelled
9.2
23
Fold: Rubredoxin-likeSuperfamily: Microbial and mitochondrial ADK, insert "zinc finger" domainFamily: Microbial and mitochondrial ADK, insert "zinc finger" domain40 c2ba0A_
not modelled
9.1
42
PDB header: rna binding proteinChain: A: PDB Molecule: archeal exosome rna binding protein rrp4;PDBTitle: archaeal exosome core
41 d2z0sa1
not modelled
7.9
56
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like42 c2gwhA_
not modelled
7.6
19
PDB header: transferaseChain: A: PDB Molecule: sulfotransferase 1c2;PDBTitle: human sulfotranferase sult1c2 in complex with pap and2 pentachlorophenol
43 c2jysA_
not modelled
7.4
20
PDB header: hydrolaseChain: A: PDB Molecule: protease/reverse transcriptase;PDBTitle: solution structure of simian foamy virus (mac) protease
44 d1okja2
not modelled
7.3
22
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: YeaZ-like45 c2ky9A_
not modelled
7.0
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein ydhk;PDBTitle: solution nmr structure of ydhk c-terminal domain from b.subtilis,2 northeast structural genomics consortium target target sr518
46 d1j99a_
not modelled
6.6
26
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase47 d1ubea2
not modelled
6.4
24
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain48 c2nn6G_
not modelled
6.4
50
PDB header: hydrolase/transferaseChain: G: PDB Molecule: exosome complex exonuclease rrp40;PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
49 d1ls6a_
not modelled
6.3
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase50 c3u3oA_
not modelled
6.3
21
PDB header: transferaseChain: A: PDB Molecule: sulfotransferase 1a1;PDBTitle: crystal structure of human sult1a1 bound to pap and two 3-cyano-7-2 hydroxycoumarin
51 d1mo6a2
not modelled
6.3
24
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain52 d1c0ma2
not modelled
6.2
12
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain53 c2dgyA_
not modelled
5.9
36
PDB header: translationChain: A: PDB Molecule: mgc11102 protein;PDBTitle: solution structure of the eukaryotic initiation factor 1a2 in mgc11102 protein
54 c2zkrq_
not modelled
5.9
15
PDB header: ribosomal protein/rnaChain: Q: PDB Molecule: rna expansion segment es31 part ii;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
55 d2a3ra1
not modelled
5.8
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase56 c1zd1B_
not modelled
5.7
19
PDB header: transferaseChain: B: PDB Molecule: sulfotransferase 4a1;PDBTitle: human sulfortransferase sult4a1
57 c3lyiA_
not modelled
5.6
14
PDB header: transcriptionChain: A: PDB Molecule: bromodomain-containing protein 1;PDBTitle: pwwp domain of human bromodomain-containing protein 1