1 c3du1X_
34.3
19
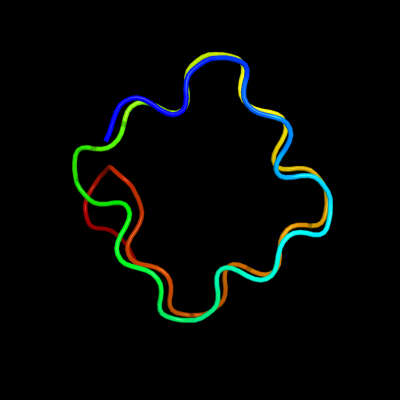
PDB header: structural proteinChain: X: PDB Molecule: all3740 protein;PDBTitle: the 2.0 angstrom resolution crystal structure of hetl, a pentapeptide2 repeat protein involved in heterocyst differentiation regulation from3 the cyanobacterium nostoc sp. strain pcc 7120
2 c3nb2B_
33.9
23
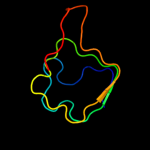
PDB header: ligaseChain: B: PDB Molecule: secreted effector protein;PDBTitle: crystal structure of e. coli o157:h7 effector protein nlel
3 c2xtwB_
18.7
12
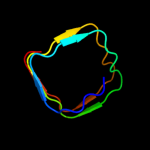
PDB header: cell cycleChain: B: PDB Molecule: qnrb1;PDBTitle: structure of qnrb1 (full length), a plasmid-mediated2 fluoroquinolone resistance protein
4 d2j8ia1
17.6
23
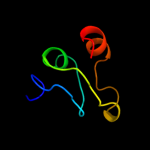
Fold: Single-stranded right-handed beta-helixSuperfamily: Pentapeptide repeat-likeFamily: Pentapeptide repeats5 c2xt4B_
11.9
18
PDB header: cell cycleChain: B: PDB Molecule: mcbg-like protein;PDBTitle: structure of the pentapeptide repeat protein albg, a2 resistance factor for the topoisomerase poison albicidin.
6 d1z0aa1
11.6
27
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins7 c2j8iB_
10.9
17
PDB header: toxinChain: B: PDB Molecule: np275;PDBTitle: structure of np275, a pentapeptide repeat protein from2 nostoc punctiforme
8 d2naca2
10.7
33
Fold: Flavodoxin-likeSuperfamily: Formate/glycerate dehydrogenase catalytic domain-likeFamily: Formate/glycerate dehydrogenases, substrate-binding domain9 c3r67A_
10.6
29
PDB header: hydrolaseChain: A: PDB Molecule: putative glycosidase;PDBTitle: crystal structure of a putative glycosidase (bt_4094) from bacteroides2 thetaiotaomicron vpi-5482 at 2.30 a resolution
10 d2j8ka1
10.4
17
Fold: Single-stranded right-handed beta-helixSuperfamily: Pentapeptide repeat-likeFamily: Pentapeptide repeats11 d1wmxa_
10.3
19
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Family 30 carbohydrate binding module, CBM30 (PKD repeat)12 d2b4wa1
10.1
50
Fold: 5-bladed beta-propellerSuperfamily: Arabinanase/levansucrase/invertaseFamily: LmjF10.1260-like13 c2o6wA_
9.6
16
PDB header: unknown functionChain: A: PDB Molecule: repeat five residue (rfr) protein orPDBTitle: crystal structure of a pentapeptide repeat protein (rfr23)2 from the cyanobacterium cyanothece 51142
14 c3pssB_
9.0
14
PDB header: cell cycleChain: B: PDB Molecule: qnr;PDBTitle: crystal structure of ahqnr, the qnr protein from aeromonas hydrophila2 (p21 crystal form)
15 c2yuiA_
8.5
21
PDB header: apoptosisChain: A: PDB Molecule: anamorsin;PDBTitle: solution structure of the n-terminal domain in human2 cytokine-induced apoptosis inhibitor anamorsin
16 c3n90A_
8.2
23
PDB header: unknown functionChain: A: PDB Molecule: thylakoid lumenal 15 kda protein 1, chloroplastic;PDBTitle: the 1.7 angstrom resolution crystal structure of at2g44920, a2 pentapeptide repeat protein from arabidopsis thaliana thylakoid3 lumen.
17 d2f3la1
8.0
18
Fold: Single-stranded right-handed beta-helixSuperfamily: Pentapeptide repeat-likeFamily: Pentapeptide repeats18 c2w7zB_
7.8
15
PDB header: inhibitorChain: B: PDB Molecule: pentapeptide repeat family protein;PDBTitle: structure of the pentapeptide repeat protein efsqnr, a dna2 gyrase inhibitor. free amines modified by cyclic3 pentylation with glutaraldehyde.
19 d2zjrj1
7.7
26
Fold: alpha/beta-HammerheadSuperfamily: Ribosomal protein L16p/L10eFamily: Ribosomal protein L16p20 d2bm5a1
7.7
12
Fold: Single-stranded right-handed beta-helixSuperfamily: Pentapeptide repeat-likeFamily: Pentapeptide repeats21 d2coba1
not modelled
7.6
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Psq domain22 c2g0yA_
not modelled
7.2
26
PDB header: unknown functionChain: A: PDB Molecule: pentapeptide repeat protein;PDBTitle: crystal structure of a lumenal pentapeptide repeat protein from2 cyanothece sp 51142 at 2.3 angstrom resolution. tetragonal crystal3 form
23 d2j01q1
not modelled
6.9
35
Fold: alpha/beta-HammerheadSuperfamily: Ribosomal protein L16p/L10eFamily: Ribosomal protein L16p24 c3e3uA_
not modelled
6.6
27
PDB header: hydrolaseChain: A: PDB Molecule: peptide deformylase;PDBTitle: crystal structure of mycobacterium tuberculosis peptide2 deformylase in complex with inhibitor
25 c3bboO_
not modelled
6.5
35
PDB header: ribosomeChain: O: PDB Molecule: ribosomal protein l16;PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
26 d2gyck1
not modelled
6.4
26
Fold: alpha/beta-HammerheadSuperfamily: Ribosomal protein L16p/L10eFamily: Ribosomal protein L16p27 d1iuqa_
not modelled
6.1
21
Fold: Glycerol-3-phosphate (1)-acyltransferaseSuperfamily: Glycerol-3-phosphate (1)-acyltransferaseFamily: Glycerol-3-phosphate (1)-acyltransferase28 c3aqoD_
not modelled
5.6
21
PDB header: membrane proteinChain: D: PDB Molecule: probable secdf protein-export membrane protein;PDBTitle: structure and function of a membrane component secdf that enhances2 protein export
29 c2k3jA_
not modelled
5.6
78
PDB header: oxidoreductaseChain: A: PDB Molecule: mitochondrial intermembrane space import andPDBTitle: the solution structure of human mia40
30 c2jx0A_
not modelled
5.6
20
PDB header: cell adhesion, signaling proteinChain: A: PDB Molecule: arf gtpase-activating protein git1;PDBTitle: the paxillin-binding domain (pbd) of g protein coupled2 receptor (gpcr)-kinase (grk) interacting protein 1 (git1)
31 d1kshb_
not modelled
5.6
38
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: RhoGDI-like32 c3nw8B_
not modelled
5.4
19
PDB header: viral proteinChain: B: PDB Molecule: envelope glycoprotein b;PDBTitle: glycoprotein b from herpes simplex virus type 1, y179s mutant, high-ph