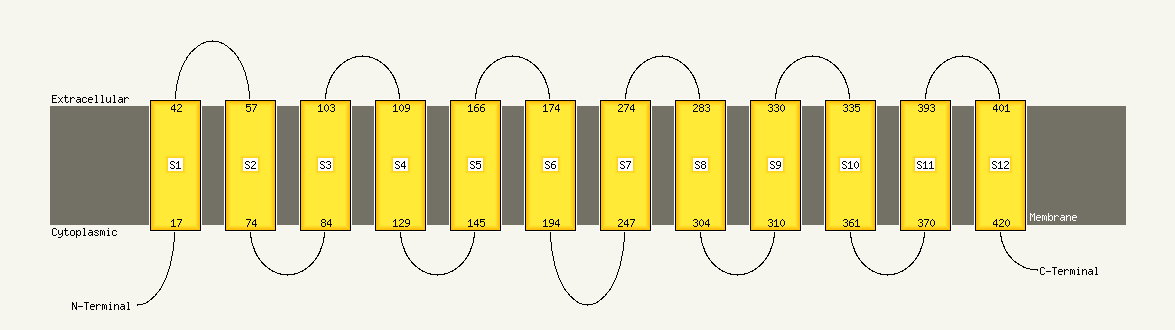
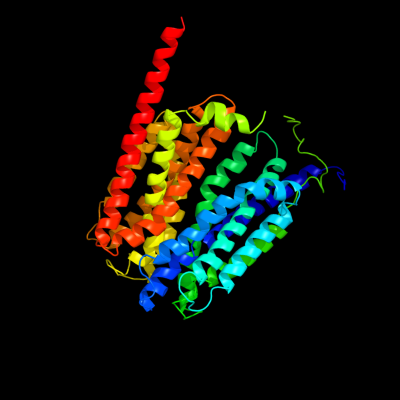
| 1 | d1pw4a_
|
|
 |
100.0 |
12 |
Fold:MFS general substrate transporter
Superfamily:MFS general substrate transporter
Family:Glycerol-3-phosphate transporter |
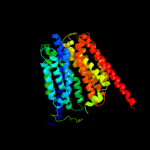
| 2 | d1pv7a_
|
|
 |
100.0 |
10 |
Fold:MFS general substrate transporter
Superfamily:MFS general substrate transporter
Family:LacY-like proton/sugar symporter |
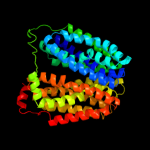
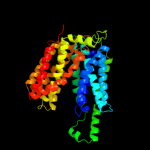
| 3 | c3o7pA_
|
|
 |
100.0 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
|
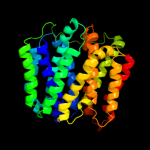
| 4 | c2gfpA_
|
|
 |
99.9 |
16 |
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
|
| 5 | c2xutC_
|
|
 |
99.9 |
13 |
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
|
| 6 | c3b9yA_
|
|
 |
70.4 |
10 |
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
|
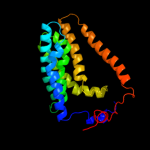
| 7 | c3hd6A_
|
|
 |
60.8 |
9 |
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
|
| 8 | c3qnqD_
|
|
 |
52.8 |
13 |
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
|
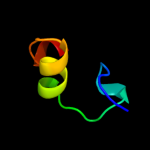
| 9 | c3pvpA_
|
|
 |
27.3 |
21 |
PDB header:dna binding protein/dna
Chain: A: PDB Molecule:chromosomal replication initiator protein dnaa;
PDBTitle: structure of mycobacterium tuberculosis dnaa-dbd in complex with box22 dna
|
| 10 | d1fs1b1
|
|
 |
26.9 |
8 |
Fold:Skp1 dimerisation domain-like
Superfamily:Skp1 dimerisation domain-like
Family:Skp1 dimerisation domain-like |
| 11 | d1fs2b1
|
|
 |
21.1 |
8 |
Fold:Skp1 dimerisation domain-like
Superfamily:Skp1 dimerisation domain-like
Family:Skp1 dimerisation domain-like |
| 12 | c3ff5B_
|
|
 |
19.9 |
12 |
PDB header:protein transport
Chain: B: PDB Molecule:peroxisomal biogenesis factor 14;
PDBTitle: crystal structure of the conserved n-terminal domain of the2 peroxisomal matrix-protein-import receptor, pex14p
|
| 13 | c2jlnA_
|
|
 |
18.0 |
10 |
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
|
| 14 | c3c9pA_
|
|
 |
17.3 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein sp1917;
PDBTitle: crystal structure of uncharacterized protein sp1917
|
| 15 | d1j1va_
|
|
 |
16.8 |
22 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:TrpR-like
Family:Chromosomal replication initiation factor DnaA C-terminal domain IV |
| 16 | d1nexa1
|
|
 |
16.2 |
0 |
Fold:Skp1 dimerisation domain-like
Superfamily:Skp1 dimerisation domain-like
Family:Skp1 dimerisation domain-like |
| 17 | c2g9pA_
|
|
 |
15.0 |
29 |
PDB header:antimicrobial protein
Chain: A: PDB Molecule:antimicrobial peptide latarcin 2a;
PDBTitle: nmr structure of a novel antimicrobial peptide, latarcin 2a,2 from spider (lachesana tarabaevi) venom
|
| 18 | d2ovra1
|
|
 |
14.5 |
8 |
Fold:Skp1 dimerisation domain-like
Superfamily:Skp1 dimerisation domain-like
Family:Skp1 dimerisation domain-like |
| 19 | c2w85A_
|
|
 |
13.0 |
10 |
PDB header:protein transport
Chain: A: PDB Molecule:peroxisomal membrane anchor protein pex14;
PDBTitle: structure of pex14 in compex with pex19
|
| 20 | c3hkzY_
|
|
 |
13.0 |
16 |
PDB header:transferase
Chain: Y: PDB Molecule:dna-directed rna polymerase subunit 13;
PDBTitle: the x-ray crystal structure of rna polymerase from archaea
|
| 21 | c2p1nD_ |
|
not modelled |
12.9 |
15 |
PDB header:signaling protein
Chain: D: PDB Molecule:skp1-like protein 1a;
PDBTitle: mechanism of auxin perception by the tir1 ubiqutin ligase
|
| 22 | d1l8qa1 |
|
not modelled |
12.9 |
7 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:TrpR-like
Family:Chromosomal replication initiation factor DnaA C-terminal domain IV |
| 23 | c2f9jP_ |
|
not modelled |
11.8 |
13 |
PDB header:rna binding protein
Chain: P: PDB Molecule:splicing factor 3b subunit 1;
PDBTitle: 3.0 angstrom resolution structure of a y22m mutant of the spliceosomal2 protein p14 bound to a region of sf3b155
|
| 24 | d1xwya1 |
|
not modelled |
9.2 |
7 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:TatD Mg-dependent DNase-like |
| 25 | d1xpja_ |
|
not modelled |
8.9 |
11 |
Fold:HAD-like
Superfamily:HAD-like
Family:Hypothetical protein VC0232 |
| 26 | c3hkzZ_ |
|
not modelled |
8.9 |
16 |
PDB header:transferase
Chain: Z: PDB Molecule:dna-directed rna polymerase subunit 13;
PDBTitle: the x-ray crystal structure of rna polymerase from archaea
|
| 27 | d1ymga1 |
|
not modelled |
8.5 |
2 |
Fold:Aquaporin-like
Superfamily:Aquaporin-like
Family:Aquaporin-like |
| 28 | c1ymgA_ |
|
not modelled |
8.5 |
2 |
PDB header:membrane protein
Chain: A: PDB Molecule:lens fiber major intrinsic protein;
PDBTitle: the channel architecture of aquaporin o at 2.2 angstrom resolution
|
| 29 | c3mmyF_ |
|
not modelled |
8.4 |
14 |
PDB header:nuclear protein
Chain: F: PDB Molecule:nuclear pore complex protein nup98;
PDBTitle: structural and functional analysis of the interaction between the2 nucleoporin nup98 and the mrna export factor rae1
|
| 30 | c3m91B_ |
|
not modelled |
8.2 |
25 |
PDB header:hydrolase regulator
Chain: B: PDB Molecule:prokaryotic ubiquitin-like protein pup;
PDBTitle: crystal structure of the prokaryotic ubiquitin-like protein (pup)2 complexed with the amino terminal coiled coil of the mycobacterium3 tuberculosis proteasomal atpase mpa
|
| 31 | d1xeqa1 |
|
not modelled |
8.2 |
40 |
Fold:S15/NS1 RNA-binding domain
Superfamily:S15/NS1 RNA-binding domain
Family:N-terminal, RNA-binding domain of nonstructural protein NS1 |
| 32 | c2vpzG_ |
|
not modelled |
7.9 |
19 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:hypothetical membrane spanning protein;
PDBTitle: polysulfide reductase native structure
|
| 33 | c2y0sJ_ |
|
not modelled |
7.7 |
11 |
PDB header:transferase
Chain: J: PDB Molecule:rna polymerase subunit 13;
PDBTitle: crystal structure of sulfolobus shibatae rna polymerase in2 p21 space group
|
| 34 | c2y0sQ_ |
|
not modelled |
7.7 |
11 |
PDB header:transferase
Chain: Q: PDB Molecule:rna polymerase subunit 13;
PDBTitle: crystal structure of sulfolobus shibatae rna polymerase in2 p21 space group
|
| 35 | c3r66A_ |
|
not modelled |
7.7 |
40 |
PDB header:viral protein/antiviral protein
Chain: A: PDB Molecule:non-structural protein 1;
PDBTitle: crystal structure of human isg15 in complex with ns1 n-terminal region2 from influenza virus b, northeast structural genomics consortium3 target ids hx6481, hr2873, and or2
|
| 36 | d2a1la1 |
|
not modelled |
7.6 |
7 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:Phoshatidylinositol transfer protein, PITP |
| 37 | d1rsob_ |
|
not modelled |
7.5 |
13 |
Fold:L27 domain
Superfamily:L27 domain
Family:L27 domain |
| 38 | c3m9dI_ |
|
not modelled |
7.2 |
20 |
PDB header:chaperone
Chain: I: PDB Molecule:prokaryotic ubiquitin-like protein pup;
PDBTitle: crystal structure of the prokaryotic ubiquintin-like protein pup2 complexed with the hexameric proteasomal atpase mpa which includes3 the amino terminal coiled coil domain and the inter domain
|
| 39 | c1egpA_ |
|
not modelled |
7.1 |
20 |
PDB header:proteinase inhibitor
Chain: A: PDB Molecule:eglin-c;
PDBTitle: proteinase inhibitor eglin c with hydrolysed reactive center
|
| 40 | c2l16A_ |
|
not modelled |
7.1 |
11 |
PDB header:protein transport
Chain: A: PDB Molecule:sec-independent protein translocase protein tatad;
PDBTitle: solution structure of bacillus subtilits tatad protein in dpc micelles
|
| 41 | c2waqQ_ |
|
not modelled |
7.0 |
11 |
PDB header:transcription
Chain: Q: PDB Molecule:dna-directed rna polymerase rpo13 subunit;
PDBTitle: the complete structure of the archaeal 13-subunit dna-2 directed rna polymerase
|
| 42 | c2rddB_ |
|
not modelled |
6.8 |
6 |
PDB header:membrane protein/transport protein
Chain: B: PDB Molecule:upf0092 membrane protein yajc;
PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
|
| 43 | d1f15a_ |
|
not modelled |
6.7 |
17 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Bromoviridae-like VP |
| 44 | d2p7vb1 |
|
not modelled |
6.7 |
6 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
| 45 | c1cirA_ |
|
not modelled |
6.7 |
17 |
PDB header:serine protease inhibitor
Chain: A: PDB Molecule:chymotrypsin inhibitor 2;
PDBTitle: complex of two fragments of ci2 [(1-40)(dot)(41-64)]
|
| 46 | d1j4na_ |
|
not modelled |
6.6 |
6 |
Fold:Aquaporin-like
Superfamily:Aquaporin-like
Family:Aquaporin-like |
| 47 | d1t27a_ |
|
not modelled |
6.6 |
7 |
Fold:TBP-like
Superfamily:Bet v1-like
Family:Phoshatidylinositol transfer protein, PITP |
| 48 | d1ofcx1 |
|
not modelled |
6.6 |
0 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Myb/SANT domain |
| 49 | c2diiA_ |
|
not modelled |
6.6 |
5 |
PDB header:transcription
Chain: A: PDB Molecule:tfiih basal transcription factor complex p62
PDBTitle: solution structure of the bsd domain of human tfiih basal2 transcription factor complex p62 subunit
|
| 50 | c1nexC_ |
|
not modelled |
6.5 |
0 |
PDB header:ligase, cell cycle
Chain: C: PDB Molecule:centromere dna-binding protein complex cbf3
PDBTitle: crystal structure of scskp1-sccdc4-cpd peptide complex
|
| 51 | d1xjsa_ |
|
not modelled |
6.5 |
7 |
Fold:SufE/NifU
Superfamily:SufE/NifU
Family:NifU/IscU domain |
| 52 | c1dgrW_ |
|
not modelled |
6.5 |
6 |
PDB header:plant protein
Chain: W: PDB Molecule:canavalin;
PDBTitle: refined crystal structure of canavalin from jack bean
|
| 53 | d2j85a1 |
|
not modelled |
6.4 |
15 |
Fold:STIV B116-like
Superfamily:STIV B116-like
Family:STIV B116-like |
| 54 | c3gg7A_ |
|
not modelled |
6.4 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized metalloprotein;
PDBTitle: crystal structure of an uncharacterized metalloprotein from2 deinococcus radiodurans
|
| 55 | d2ppxa1 |
|
not modelled |
6.4 |
4 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:SinR domain-like |
| 56 | c2ppxA_ |
|
not modelled |
6.4 |
4 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein atu1735;
PDBTitle: crystal structure of a hth xre-family like protein from agrobacterium2 tumefaciens
|
| 57 | c2kz6A_ |
|
not modelled |
6.3 |
29 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of protein cv0426 from chromobacterium violaceum,2 northeast structural genomics consortium (nesg) target cvt2
|
| 58 | d2ocda1 |
|
not modelled |
6.3 |
5 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 59 | d1jmxa1 |
|
not modelled |
6.2 |
8 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:Quinohemoprotein amine dehydrogenase A chain, domains 1 and 2 |
| 60 | d2diia1 |
|
not modelled |
6.2 |
5 |
Fold:BSD domain-like
Superfamily:BSD domain-like
Family:BSD domain |
| 61 | d1j6oa_ |
|
not modelled |
6.1 |
7 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:TatD Mg-dependent DNase-like |
| 62 | c3h00A_ |
|
not modelled |
6.1 |
20 |
PDB header:viral protein
Chain: A: PDB Molecule:envelope glycoprotein gp160;
PDBTitle: structure of the c-terminal domain of a putative hiv-1 gp412 fusion intermediate
|
| 63 | c2kdcC_ |
|
not modelled |
5.8 |
11 |
PDB header:transferase
Chain: C: PDB Molecule:diacylglycerol kinase;
PDBTitle: nmr solution structure of e. coli diacylglycerol kinase2 (dagk) in dpc micelles
|
| 64 | c2wb1Q_ |
|
not modelled |
5.6 |
11 |
PDB header:transcription
Chain: Q: PDB Molecule:dna-directed rna polymerase rpo13 subunit;
PDBTitle: the complete structure of the archaeal 13-subunit dna-2 directed rna polymerase
|
| 65 | c3bs3A_ |
|
not modelled |
5.6 |
17 |
PDB header:dna binding protein
Chain: A: PDB Molecule:putative dna-binding protein;
PDBTitle: crystal structure of a putative dna-binding protein from bacteroides2 fragilis
|
| 66 | d1agxa_ |
|
not modelled |
5.6 |
20 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 67 | d1pbya1 |
|
not modelled |
5.5 |
8 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:Quinohemoprotein amine dehydrogenase A chain, domains 1 and 2 |
| 68 | c3rkoK_ |
|
not modelled |
5.4 |
14 |
PDB header:oxidoreductase
Chain: K: PDB Molecule:nadh-quinone oxidoreductase subunit k;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
|
| 69 | c2wwbA_ |
|
not modelled |
5.4 |
11 |
PDB header:ribosome
Chain: A: PDB Molecule:protein transport protein sec61 subunit alpha isoform 1;
PDBTitle: cryo-em structure of the mammalian sec61 complex bound to the2 actively translating wheat germ 80s ribosome
|
| 70 | c2ci2I_ |
|
not modelled |
5.3 |
17 |
PDB header:proteinase inhibitor (chymotrypsin)
Chain: I: PDB Molecule:chymotrypsin inhibitor 2;
PDBTitle: crystal and molecular structure of the serine proteinase2 inhibitor ci-2 from barley seeds
|
| 71 | d1vola2 |
|
not modelled |
5.2 |
6 |
Fold:Cyclin-like
Superfamily:Cyclin-like
Family:Transcription factor IIB (TFIIB), core domain |
| 72 | c2wb1J_ |
|
not modelled |
5.2 |
11 |
PDB header:transcription
Chain: J: PDB Molecule:dna-directed rna polymerase rpo13 subunit;
PDBTitle: the complete structure of the archaeal 13-subunit dna-2 directed rna polymerase
|
| 73 | d1zzma1 |
|
not modelled |
5.2 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:TatD Mg-dependent DNase-like |
| 74 | d1to2i_ |
|
not modelled |
5.1 |
17 |
Fold:CI-2 family of serine protease inhibitors
Superfamily:CI-2 family of serine protease inhibitors
Family:CI-2 family of serine protease inhibitors |
| 75 | d1csei_ |
|
not modelled |
5.1 |
17 |
Fold:CI-2 family of serine protease inhibitors
Superfamily:CI-2 family of serine protease inhibitors
Family:CI-2 family of serine protease inhibitors |
| 76 | d1ypci_ |
|
not modelled |
5.1 |
21 |
Fold:CI-2 family of serine protease inhibitors
Superfamily:CI-2 family of serine protease inhibitors
Family:CI-2 family of serine protease inhibitors |