| 1 |
|
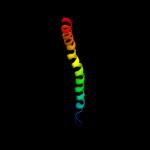
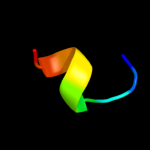
PDB 1e8o chain A
Region: 3 - 13
Aligned: 11
Modelled: 11
Confidence: 36.4%
Identity: 36%
Fold: Signal recognition particle alu RNA binding heterodimer, SRP9/14
Superfamily: Signal recognition particle alu RNA binding heterodimer, SRP9/14
Family: Signal recognition particle alu RNA binding heterodimer, SRP9/14
Phyre2
| 2 |
|
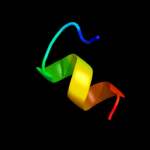
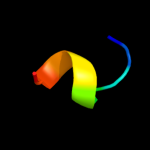
PDB 2knc chain B
Region: 13 - 64
Aligned: 52
Modelled: 52
Confidence: 18.0%
Identity: 19%
PDB header:cell adhesion
Chain: B: PDB Molecule:integrin beta-3;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 3 |
|
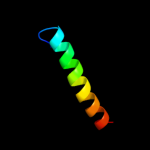
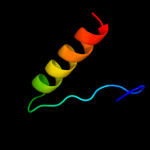
PDB 1q88 chain B
Region: 4 - 15
Aligned: 12
Modelled: 12
Confidence: 15.2%
Identity: 33%
PDB header:dna binding protein
Chain: B: PDB Molecule:39 kda initiator binding protein;
PDBTitle: crystal structure of the c-domain of the t.vaginalis inr2 binding protein, ibp39 (monoclinic form)
Phyre2
| 4 |
|
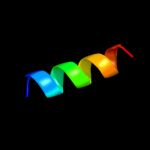
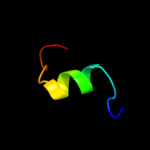
PDB 1q88 chain A
Region: 4 - 14
Aligned: 11
Modelled: 11
Confidence: 13.1%
Identity: 36%
Fold: 39 kda initiator binding protein, IBP39, C-terminal domains
Superfamily: 39 kda initiator binding protein, IBP39, C-terminal domains
Family: 39 kda initiator binding protein, IBP39, C-terminal domains
Phyre2
| 5 |
|
PDB 2fv2 chain A
Region: 18 - 32
Aligned: 15
Modelled: 15
Confidence: 12.2%
Identity: 20%
PDB header:transcription
Chain: A: PDB Molecule:rcd1 required for cell differentiation1 homolog;
PDBTitle: crystal structure analysis of human rcd-1 conserved region
Phyre2
| 6 |
|
PDB 1nek chain D
Region: 13 - 40
Aligned: 28
Modelled: 28
Confidence: 12.1%
Identity: 25%
Fold: Heme-binding four-helical bundle
Superfamily: Fumarate reductase respiratory complex transmembrane subunits
Family: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)
Phyre2
| 7 |
|
PDB 3hlz chain A
Region: 22 - 35
Aligned: 14
Modelled: 14
Confidence: 10.1%
Identity: 36%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein bt_1490;
PDBTitle: crystal structure of bt_1490 (np_810393.1) from bacteroides2 thetaiotaomicron vpi-5482 at 1.50 a resolution
Phyre2
| 8 |
|
PDB 1914 chain A domain 1
Region: 5 - 13
Aligned: 9
Modelled: 9
Confidence: 9.3%
Identity: 44%
Fold: Signal recognition particle alu RNA binding heterodimer, SRP9/14
Superfamily: Signal recognition particle alu RNA binding heterodimer, SRP9/14
Family: Signal recognition particle alu RNA binding heterodimer, SRP9/14
Phyre2
| 9 |
|
PDB 1s5r chain A
Region: 10 - 26
Aligned: 17
Modelled: 17
Confidence: 8.5%
Identity: 24%
PDB header:transcription
Chain: A: PDB Molecule:high mobility group box transcription factor 1;
PDBTitle: solution structure of hbp1 sid-msin3a pah2 complex
Phyre2
| 10 |
|
PDB 1914 chain A
Region: 5 - 13
Aligned: 9
Modelled: 9
Confidence: 8.1%
Identity: 44%
PDB header:alu domain
Chain: A: PDB Molecule:signal recognition particle 9/14 fusion protein;
PDBTitle: signal recognition particle alu rna binding heterodimer, srp9/14
Phyre2
| 11 |
|
PDB 3m4r chain A
Region: 14 - 42
Aligned: 29
Modelled: 29
Confidence: 8.0%
Identity: 3%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: structure of the n-terminal class ii aldolase domain of a conserved2 protein from thermoplasma acidophilum
Phyre2
| 12 |
|
PDB 3s8m chain A
Region: 3 - 23
Aligned: 21
Modelled: 21
Confidence: 7.6%
Identity: 19%
PDB header:oxidoreductase
Chain: A: PDB Molecule:enoyl-acp reductase;
PDBTitle: the crystal structure of fabv
Phyre2
| 13 |
|
PDB 2cq8 chain A
Region: 10 - 17
Aligned: 8
Modelled: 8
Confidence: 6.4%
Identity: 38%
PDB header:oxidoreductase
Chain: A: PDB Molecule:10-formyltetrahydrofolate dehydrogenase;
PDBTitle: solution structure of rsgi ruh-033, a pp-binding domain of2 10-fthfdh from human cdna
Phyre2
| 14 |
|
PDB 2je6 chain I domain 2
Region: 14 - 21
Aligned: 8
Modelled: 8
Confidence: 5.9%
Identity: 0%
Fold: Barrel-sandwich hybrid
Superfamily: Ribosomal L27 protein-like
Family: ECR1 N-terminal domain-like
Phyre2
| 15 |
|
PDB 2fk5 chain B
Region: 14 - 52
Aligned: 39
Modelled: 39
Confidence: 5.7%
Identity: 13%
PDB header:lyase
Chain: B: PDB Molecule:fuculose-1-phosphate aldolase;
PDBTitle: crystal structure of l-fuculose-1-phosphate aldolase from thermus2 thermophilus hb8
Phyre2
| 16 |
|
PDB 1sgg chain A
Region: 3 - 21
Aligned: 19
Modelled: 19
Confidence: 5.6%
Identity: 11%
Fold: SAM domain-like
Superfamily: SAM/Pointed domain
Family: SAM (sterile alpha motif) domain
Phyre2