1 c3k5bE_
98.5
14
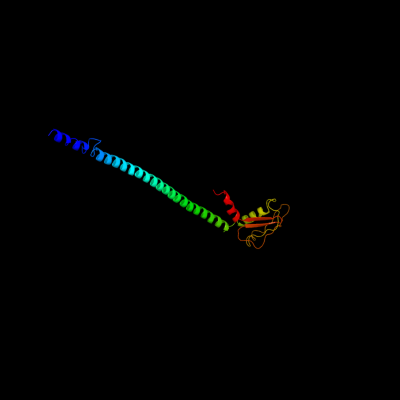
PDB header: hydrolaseChain: E: PDB Molecule: v-type atp synthase subunit e;PDBTitle: crystal structure of the peripheral stalk of thermus thermophilus h+-2 atpase/synthase
2 d2dm9a1
98.3
13
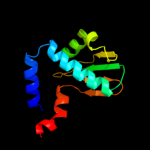
Fold: FwdE/GAPDH domain-likeSuperfamily: V-type ATPase subunit E-likeFamily: V-type ATPase subunit E3 c2dm9B_
98.3
13
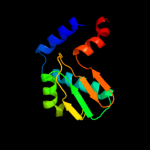
PDB header: hydrolaseChain: B: PDB Molecule: v-type atp synthase subunit e;PDBTitle: crystal structure of ph1978 from pyrococcus horikoshii ot3
4 c3lg8B_
97.0
22
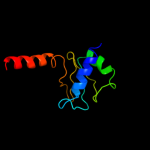
PDB header: hydrolaseChain: B: PDB Molecule: a-type atp synthase subunit e;PDBTitle: crystal structure of the c-terminal part of subunit e (e101-206) from2 methanocaldococcus jannaschii of a1ao atp synthase
5 c2lc0A_
90.9
18
PDB header: protein bindingChain: A: PDB Molecule: putative uncharacterized protein tb39.8;PDBTitle: rv0020c_nter structure
6 c2c4rL_
53.6
17
PDB header: hydrolaseChain: L: PDB Molecule: ribonuclease e;PDBTitle: catalytic domain of e. coli rnase e
7 c3sftA_
23.6
12
PDB header: hydrolaseChain: A: PDB Molecule: chemotaxis response regulator protein-glutamatePDBTitle: crystal structure of thermotoga maritima cheb methylesterase catalytic2 domain
8 c2rrlA_
22.6
20
PDB header: protein transportChain: A: PDB Molecule: flagellar hook-length control protein;PDBTitle: solution structure of the c-terminal domain of the flik
9 d3bzka2
21.1
29
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Tex HhH-containing domain-like10 d1y5ia1
20.1
16
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain11 d1zy9a1
17.1
22
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: YicI N-terminal domain-like12 c2e0zA_
16.7
10
PDB header: virus like particleChain: A: PDB Molecule: virus-like particle;PDBTitle: crystal structure of virus-like particle from pyrococcus2 furiosus
13 d2jioa1
16.5
13
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain14 d1p9qc3
15.8
13
Fold: Ferredoxin-likeSuperfamily: EF-G C-terminal domain-likeFamily: Hypothetical protein AF0491, C-terminal domain15 c2a45J_
14.1
18
PDB header: hydrolase/hydrolase inhibitorChain: J: PDB Molecule: fibrinogen alpha chain;PDBTitle: crystal structure of the complex between thrombin and the central "e"2 region of fibrin
16 d1t95a3
12.7
13
Fold: Ferredoxin-likeSuperfamily: EF-G C-terminal domain-likeFamily: Hypothetical protein AF0491, C-terminal domain17 c3dktD_
12.6
17
PDB header: structural protein/virus like particleChain: D: PDB Molecule: maritimacin;PDBTitle: crystal structure of thermotoga maritima encapsulin
18 c3tr6A_
12.3
18
PDB header: transferaseChain: A: PDB Molecule: o-methyltransferase;PDBTitle: structure of a o-methyltransferase from coxiella burnetii
19 c3u7jA_
11.5
21
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: crystal structure of ribose-5-phosphate isomerase a from burkholderia2 thailandensis
20 d2iv2x1
11.2
16
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain21 d1pkxa1
not modelled
10.2
10
Fold: Methylglyoxal synthase-likeSuperfamily: Methylglyoxal synthase-likeFamily: Inosicase22 d2q22a1
not modelled
8.8
12
Fold: Ava3019-likeSuperfamily: Ava3019-likeFamily: Ava3019-like23 c3h9xB_
not modelled
8.2
12
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein pspto_3016;PDBTitle: crystal structure of the pspto_3016 protein from2 pseudomonas syringae, northeast structural genomics3 consortium target psr293
24 c3cagF_
not modelled
7.7
16
PDB header: dna binding proteinChain: F: PDB Molecule: arginine repressor;PDBTitle: crystal structure of the oligomerization domain hexamer of the2 arginine repressor protein from mycobacterium tuberculosis in complex3 with 9 arginines.
25 d2b0ja2
not modelled
7.7
13
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain26 c2ki8A_
not modelled
7.5
10
PDB header: oxidoreductaseChain: A: PDB Molecule: tungsten formylmethanofuran dehydrogenase,PDBTitle: solution nmr structure of tungsten formylmethanofuran2 dehydrogenase subunit d from archaeoglobus fulgidus,3 northeast structural genomics consortium target att7
27 c3cbnA_
not modelled
7.4
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved protein mth639;PDBTitle: crystal structure of a conserved protein (mth639) from2 methanobacterium thermoautotrophicum
28 d1uv7a_
not modelled
7.3
12
Fold: RRF/tRNA synthetase additional domain-likeSuperfamily: General secretion pathway protein M, EpsMFamily: General secretion pathway protein M, EpsM29 c1uv7A_
not modelled
7.3
12
PDB header: transportChain: A: PDB Molecule: general secretion pathway protein m;PDBTitle: periplasmic domain of epsm from vibrio cholerae
30 d1go3e2
not modelled
7.1
21
Fold: Dodecin subunit-likeSuperfamily: N-terminal, heterodimerisation domain of RBP7 (RpoE)Family: N-terminal, heterodimerisation domain of RBP7 (RpoE)31 d2c35b2
not modelled
7.1
17
Fold: Dodecin subunit-likeSuperfamily: N-terminal, heterodimerisation domain of RBP7 (RpoE)Family: N-terminal, heterodimerisation domain of RBP7 (RpoE)32 c3ebrA_
not modelled
6.9
30
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized rmlc-like cupin;PDBTitle: crystal structure of an rmlc-like cupin protein (reut_a0381) from2 ralstonia eutropha jmp134 at 2.60 a resolution
33 c3sb1B_
not modelled
6.8
23
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hydrogenase expression protein;PDBTitle: hydrogenase expression protein huph from thiobacillus denitrificans2 atcc 25259
34 c3ereD_
not modelled
6.3
14
PDB header: dna binding protein/dnaChain: D: PDB Molecule: arginine repressor;PDBTitle: crystal structure of the arginine repressor protein from mycobacterium2 tuberculosis in complex with the dna operator
35 d1chda_
not modelled
5.9
17
Fold: Methylesterase CheB, C-terminal domainSuperfamily: Methylesterase CheB, C-terminal domainFamily: Methylesterase CheB, C-terminal domain36 d1dmra1
not modelled
5.8
18
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain37 c3a6mB_
not modelled
5.8
20
PDB header: chaperoneChain: B: PDB Molecule: protein grpe;PDBTitle: crystal structure of grpe from thermus thermophilus hb8
38 d1tmoa1
not modelled
5.6
12
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain39 d1l2pa_
not modelled
5.5
11
Fold: Single transmembrane helixSuperfamily: F1F0 ATP synthase subunit B, membrane domainFamily: F1F0 ATP synthase subunit B, membrane domain40 d1ogya1
not modelled
5.4
17
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain41 d1ou0a_
not modelled
5.2
13
Fold: Flavodoxin-likeSuperfamily: Precorrin-8X methylmutase CbiC/CobHFamily: Precorrin-8X methylmutase CbiC/CobH42 d1cwpa_
not modelled
5.1
13
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Bromoviridae-like VP43 d1a8ya3
not modelled
5.1
14
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Calsequestrin44 d1g8ma1
not modelled
5.1
10
Fold: Methylglyoxal synthase-likeSuperfamily: Methylglyoxal synthase-likeFamily: Inosicase45 c3cxbA_
not modelled
5.1
22
PDB header: signaling proteinChain: A: PDB Molecule: protein sifa;PDBTitle: crystal structure of sifa and skip