| 1 |
|
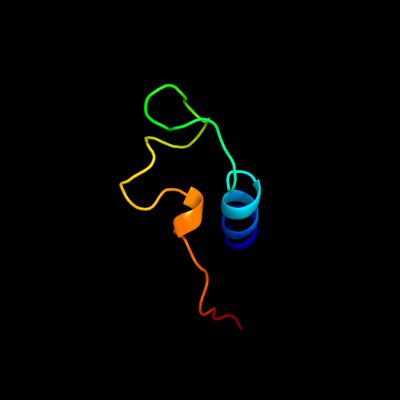
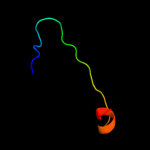
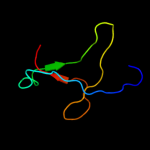
PDB 1gr0 chain A domain 1
Region: 39 - 83
Aligned: 43
Modelled: 45
Confidence: 29.5%
Identity: 23%
Fold: NAD(P)-binding Rossmann-fold domains
Superfamily: NAD(P)-binding Rossmann-fold domains
Family: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
Phyre2
| 2 |
|
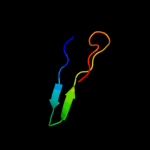
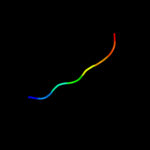
PDB 1uwc chain A
Region: 105 - 141
Aligned: 37
Modelled: 37
Confidence: 14.1%
Identity: 22%
Fold: alpha/beta-Hydrolases
Superfamily: alpha/beta-Hydrolases
Family: Fungal lipases
Phyre2
| 3 |
|
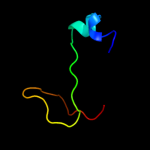
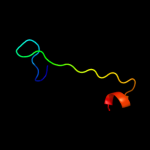
PDB 1mkf chain A
Region: 19 - 43
Aligned: 24
Modelled: 25
Confidence: 13.5%
Identity: 33%
Fold: Viral chemokine binding protein m3
Superfamily: Viral chemokine binding protein m3
Family: Viral chemokine binding protein m3
Phyre2
| 4 |
|
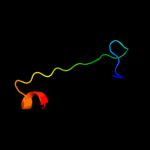
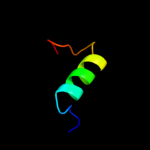
PDB 1oe4 chain A
Region: 26 - 46
Aligned: 21
Modelled: 21
Confidence: 12.8%
Identity: 29%
Fold: Uracil-DNA glycosylase-like
Superfamily: Uracil-DNA glycosylase-like
Family: Single-strand selective monofunctional uracil-DNA glycosylase SMUG1
Phyre2
| 5 |
|
PDB 3dml chain A
Region: 50 - 86
Aligned: 37
Modelled: 37
Confidence: 11.5%
Identity: 11%
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the periplasmic thioredoxin soxs from2 paracoccus pantotrophus (reduced form)
Phyre2
| 6 |
|
PDB 3ofe chain B
Region: 64 - 76
Aligned: 13
Modelled: 12
Confidence: 10.0%
Identity: 38%
PDB header:chaperone
Chain: B: PDB Molecule:ldlr chaperone boca;
PDBTitle: structured domain of drosophila melanogaster boca p41 2 2 crystal form
Phyre2
| 7 |
|
PDB 2qn6 chain A domain 1
Region: 68 - 84
Aligned: 17
Modelled: 17
Confidence: 8.9%
Identity: 41%
Fold: Reductase/isomerase/elongation factor common domain
Superfamily: Translation proteins
Family: Elongation factors
Phyre2
| 8 |
|
PDB 2yr1 chain B
Region: 54 - 79
Aligned: 26
Modelled: 26
Confidence: 8.7%
Identity: 35%
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of 3-dehydroquinate dehydratase from geobacillus2 kaustophilus hta426
Phyre2
| 9 |
|
PDB 1gqn chain A
Region: 54 - 79
Aligned: 26
Modelled: 26
Confidence: 8.2%
Identity: 31%
Fold: TIM beta/alpha-barrel
Superfamily: Aldolase
Family: Class I aldolase
Phyre2
| 10 |
|
PDB 2eoy chain A
Region: 7 - 14
Aligned: 8
Modelled: 8
Confidence: 7.7%
Identity: 63%
PDB header:transcription
Chain: A: PDB Molecule:zinc finger protein 473;
PDBTitle: solution structure of the c2h2 type zinc finger (region 557-2 589) of human zinc finger protein 473
Phyre2
| 11 |
|
PDB 2p6b chain A domain 1
Region: 107 - 125
Aligned: 19
Modelled: 19
Confidence: 7.0%
Identity: 26%
Fold: Metallo-dependent phosphatases
Superfamily: Metallo-dependent phosphatases
Family: Protein serine/threonine phosphatase
Phyre2
| 12 |
|
PDB 2p6b chain C
Region: 107 - 125
Aligned: 19
Modelled: 19
Confidence: 7.0%
Identity: 26%
PDB header:hydrolase/hydrolase regulator
Chain: C: PDB Molecule:calmodulin-dependent calcineurin a subunit alpha
PDBTitle: crystal structure of human calcineurin in complex with2 pvivit peptide
Phyre2
| 13 |
|
PDB 2exu chain A
Region: 126 - 141
Aligned: 16
Modelled: 16
Confidence: 6.4%
Identity: 25%
PDB header:transcription
Chain: A: PDB Molecule:transcription initiation protein spt4/spt5;
PDBTitle: crystal structure of saccharomyces cerevisiae transcription elongation2 factors spt4-spt5ngn domain
Phyre2
| 14 |
|
PDB 3js3 chain C
Region: 53 - 79
Aligned: 27
Modelled: 27
Confidence: 6.3%
Identity: 22%
PDB header:lyase
Chain: C: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of type i 3-dehydroquinate dehydratase (arod) from2 clostridium difficile with covalent reaction intermediate
Phyre2
| 15 |
|
PDB 1zu1 chain A domain 1
Region: 88 - 113
Aligned: 26
Modelled: 26
Confidence: 6.2%
Identity: 23%
Fold: beta-beta-alpha zinc fingers
Superfamily: beta-beta-alpha zinc fingers
Family: HkH motif-containing C2H2 finger
Phyre2
| 16 |
|
PDB 2jog chain A
Region: 107 - 125
Aligned: 19
Modelled: 19
Confidence: 5.6%
Identity: 26%
PDB header:hydrolase
Chain: A: PDB Molecule:calmodulin-dependent calcineurin a subunit alpha
PDBTitle: structure of the calcineurin-nfat complex
Phyre2
| 17 |
|
PDB 2l3i chain A
Region: 16 - 22
Aligned: 7
Modelled: 7
Confidence: 5.5%
Identity: 71%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:aoxki4a, antimicrobial peptide in spider venom;
PDBTitle: oxki4a, spider derived antimicrobial peptide
Phyre2
| 18 |
|
PDB 3h7h chain B
Region: 126 - 141
Aligned: 16
Modelled: 16
Confidence: 5.3%
Identity: 31%
PDB header:transcription
Chain: B: PDB Molecule:transcription elongation factor spt5;
PDBTitle: crystal structure of the human transcription elongation factor dsif,2 hspt4/hspt5 (176-273)
Phyre2
| 19 |
|
PDB 4a56 chain A
Region: 2 - 10
Aligned: 9
Modelled: 9
Confidence: 5.3%
Identity: 56%
PDB header:protein transport
Chain: A: PDB Molecule:pullulanase secretion protein puls;
PDBTitle: crystal structure of the type 2 secretion system pilotin2 from klebsiella oxytoca
Phyre2
| 20 |
|
PDB 3n2o chain A
Region: 30 - 92
Aligned: 51
Modelled: 51
Confidence: 5.2%
Identity: 22%
PDB header:lyase
Chain: A: PDB Molecule:biosynthetic arginine decarboxylase;
PDBTitle: x-ray crystal structure of arginine decarboxylase complexed with2 arginine from vibrio vulnificus
Phyre2
| 21 |
|
| 22 |
|