| 1 |
|
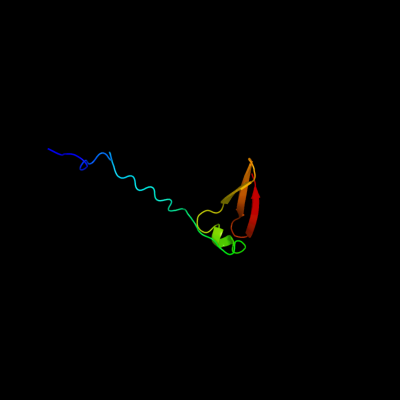
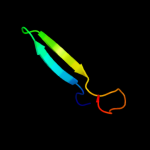
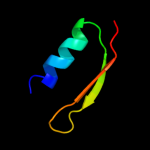
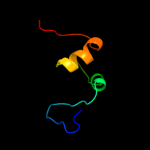
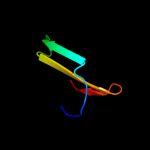
PDB 2jra chain B
Region: 1 - 63
Aligned: 63
Modelled: 63
Confidence: 99.8%
Identity: 32%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:protein rpa2121;
PDBTitle: a novel domain-swapped solution nmr structure of protein rpa2121 from2 rhodopseudomonas palustris. northeast structural genomics target rpt6
Phyre2
| 2 |
|
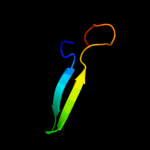
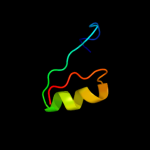
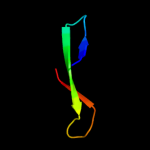
PDB 1t1u chain A domain 1
Region: 36 - 60
Aligned: 25
Modelled: 25
Confidence: 29.5%
Identity: 16%
Fold: CoA-dependent acyltransferases
Superfamily: CoA-dependent acyltransferases
Family: Choline/Carnitine O-acyltransferase
Phyre2
| 3 |
|
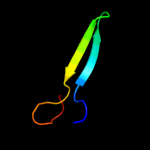
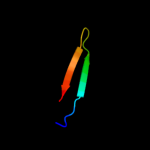
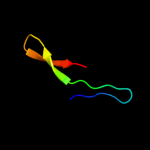
PDB 1xl7 chain A domain 1
Region: 35 - 60
Aligned: 26
Modelled: 26
Confidence: 28.7%
Identity: 12%
Fold: CoA-dependent acyltransferases
Superfamily: CoA-dependent acyltransferases
Family: Choline/Carnitine O-acyltransferase
Phyre2
| 4 |
|
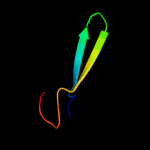
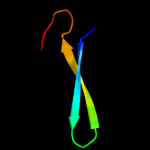
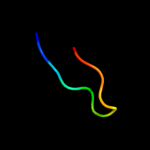
PDB 1qqr chain A
Region: 1 - 51
Aligned: 51
Modelled: 51
Confidence: 22.7%
Identity: 20%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: Staphylokinase/streptokinase
Family: Staphylokinase/streptokinase
Phyre2
| 5 |
|
PDB 1q6x chain A
Region: 36 - 60
Aligned: 25
Modelled: 25
Confidence: 17.4%
Identity: 16%
PDB header:transferase
Chain: A: PDB Molecule:choline o-acetyltransferase;
PDBTitle: crystal structure of rat choline acetyltransferase
Phyre2
| 6 |
|
PDB 1nm8 chain A domain 1
Region: 35 - 62
Aligned: 28
Modelled: 28
Confidence: 14.4%
Identity: 21%
Fold: CoA-dependent acyltransferases
Superfamily: CoA-dependent acyltransferases
Family: Choline/Carnitine O-acyltransferase
Phyre2
| 7 |
|
PDB 1t7q chain A
Region: 35 - 62
Aligned: 28
Modelled: 28
Confidence: 12.7%
Identity: 21%
PDB header:transferase
Chain: A: PDB Molecule:carnitine acetyltransferase;
PDBTitle: crystal structure of the f565a mutant of murine carnitine2 acetyltransferase in complex with carnitine and coa
Phyre2
| 8 |
|
PDB 1ndb chain A domain 1
Region: 35 - 62
Aligned: 28
Modelled: 28
Confidence: 12.3%
Identity: 21%
Fold: CoA-dependent acyltransferases
Superfamily: CoA-dependent acyltransferases
Family: Choline/Carnitine O-acyltransferase
Phyre2
| 9 |
|
PDB 1xl8 chain B
Region: 35 - 60
Aligned: 26
Modelled: 26
Confidence: 10.0%
Identity: 12%
PDB header:transferase
Chain: B: PDB Molecule:peroxisomal carnitine o-octanoyltransferase;
PDBTitle: crystal structure of mouse carnitine octanoyltransferase in2 complex with octanoylcarnitine
Phyre2
| 10 |
|
PDB 1dcj chain A
Region: 22 - 57
Aligned: 36
Modelled: 36
Confidence: 9.8%
Identity: 19%
Fold: IF3-like
Superfamily: SirA-like
Family: SirA-like
Phyre2
| 11 |
|
PDB 3tq8 chain A
Region: 32 - 63
Aligned: 32
Modelled: 32
Confidence: 9.6%
Identity: 22%
PDB header:oxidoreductase/oxidoreductase inhibitor
Chain: A: PDB Molecule:dihydrofolate reductase;
PDBTitle: structure of the dihydrofolate reductase (fola) from coxiella burnetii2 in complex with trimethoprim
Phyre2
| 12 |
|
PDB 3lyd chain A
Region: 33 - 54
Aligned: 22
Modelled: 22
Confidence: 9.3%
Identity: 9%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of putative uncharacterized protein from jonesia2 denitrificans
Phyre2
| 13 |
|
PDB 2fy2 chain A
Region: 36 - 60
Aligned: 25
Modelled: 25
Confidence: 9.0%
Identity: 16%
PDB header:transferase
Chain: A: PDB Molecule:choline o-acetyltransferase;
PDBTitle: structures of ligand bound human choline acetyltransferase2 provide insight into regulation of acetylcholine synthesis
Phyre2
| 14 |
|
PDB 3f0u chain X
Region: 14 - 63
Aligned: 50
Modelled: 50
Confidence: 8.5%
Identity: 10%
PDB header:oxidoreductase
Chain: X: PDB Molecule:trimethoprim-sensitive dihydrofolate reductase;
PDBTitle: staphylococcus aureus f98y mutant dihydrofolate reductase2 complexed with nadph and 2,4-diamino-5-[3-(3-methoxy-5-3 phenylphenyl)but-1-ynyl]-6-methylpyrimidine
Phyre2
| 15 |
|
PDB 1kv7 chain A domain 2
Region: 33 - 57
Aligned: 25
Modelled: 25
Confidence: 8.1%
Identity: 12%
Fold: Cupredoxin-like
Superfamily: Cupredoxins
Family: Multidomain cupredoxins
Phyre2
| 16 |
|
PDB 2xzm chain W
Region: 39 - 63
Aligned: 25
Modelled: 25
Confidence: 8.1%
Identity: 16%
PDB header:ribosome
Chain: W: PDB Molecule:40s ribosomal protein s4;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
Phyre2
| 17 |
|
PDB 3iz6 chain D
Region: 39 - 63
Aligned: 25
Modelled: 25
Confidence: 7.5%
Identity: 24%
PDB header:ribosome
Chain: D: PDB Molecule:40s ribosomal protein s4 (s4e);
PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
Phyre2
| 18 |
|
PDB 2zjr chain 3 domain 1
Region: 50 - 60
Aligned: 11
Modelled: 11
Confidence: 7.0%
Identity: 27%
Fold: L35p-like
Superfamily: L35p-like
Family: Ribosomal protein L35p
Phyre2
| 19 |
|
PDB 1v10 chain A domain 2
Region: 35 - 57
Aligned: 23
Modelled: 23
Confidence: 6.5%
Identity: 13%
Fold: Cupredoxin-like
Superfamily: Cupredoxins
Family: Multidomain cupredoxins
Phyre2
| 20 |
|
PDB 2hsi chain B
Region: 21 - 51
Aligned: 31
Modelled: 31
Confidence: 5.9%
Identity: 13%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative peptidase m23;
PDBTitle: crystal structure of putative peptidase m23 from2 pseudomonas aeruginosa, new york structural genomics3 consortium
Phyre2
| 21 |
|
| 22 |
|