| 1 |
|
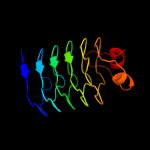
PDB 3du1 chain X
Region: 139 - 370
Aligned: 230
Modelled: 232
Confidence: 100.0%
Identity: 16%
PDB header:structural protein
Chain: X: PDB Molecule:all3740 protein;
PDBTitle: the 2.0 angstrom resolution crystal structure of hetl, a pentapeptide2 repeat protein involved in heterocyst differentiation regulation from3 the cyanobacterium nostoc sp. strain pcc 7120
Phyre2
| 2 |
|
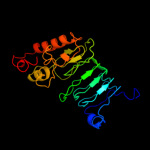
PDB 2xt4 chain B
Region: 149 - 340
Aligned: 192
Modelled: 192
Confidence: 100.0%
Identity: 15%
PDB header:cell cycle
Chain: B: PDB Molecule:mcbg-like protein;
PDBTitle: structure of the pentapeptide repeat protein albg, a2 resistance factor for the topoisomerase poison albicidin.
Phyre2
| 3 |
|
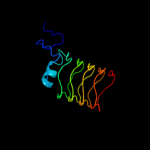
PDB 3pss chain B
Region: 164 - 370
Aligned: 206
Modelled: 207
Confidence: 100.0%
Identity: 13%
PDB header:cell cycle
Chain: B: PDB Molecule:qnr;
PDBTitle: crystal structure of ahqnr, the qnr protein from aeromonas hydrophila2 (p21 crystal form)
Phyre2
| 4 |
|
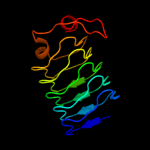
PDB 2xtw chain B
Region: 151 - 359
Aligned: 208
Modelled: 209
Confidence: 100.0%
Identity: 14%
PDB header:cell cycle
Chain: B: PDB Molecule:qnrb1;
PDBTitle: structure of qnrb1 (full length), a plasmid-mediated2 fluoroquinolone resistance protein
Phyre2
| 5 |
|
PDB 2xt4 chain A
Region: 153 - 359
Aligned: 182
Modelled: 182
Confidence: 100.0%
Identity: 16%
PDB header:cell cycle
Chain: A: PDB Molecule:mcbg-like protein;
PDBTitle: structure of the pentapeptide repeat protein albg, a2 resistance factor for the topoisomerase poison albicidin.
Phyre2
| 6 |
|
PDB 3nb2 chain B
Region: 145 - 371
Aligned: 176
Modelled: 176
Confidence: 100.0%
Identity: 14%
PDB header:ligase
Chain: B: PDB Molecule:secreted effector protein;
PDBTitle: crystal structure of e. coli o157:h7 effector protein nlel
Phyre2
| 7 |
|
PDB 2j8k chain A
Region: 151 - 326
Aligned: 176
Modelled: 176
Confidence: 99.9%
Identity: 19%
PDB header:toxin
Chain: A: PDB Molecule:np275-np276;
PDBTitle: structure of the fusion of np275 and np276, pentapeptide2 repeat proteins from nostoc punctiforme
Phyre2
| 8 |
|
PDB 2bm5 chain A domain 1
Region: 158 - 371
Aligned: 179
Modelled: 179
Confidence: 99.9%
Identity: 15%
Fold: Single-stranded right-handed beta-helix
Superfamily: Pentapeptide repeat-like
Family: Pentapeptide repeats
Phyre2
| 9 |
|
PDB 2w7z chain B
Region: 164 - 360
Aligned: 187
Modelled: 197
Confidence: 99.9%
Identity: 18%
PDB header:inhibitor
Chain: B: PDB Molecule:pentapeptide repeat family protein;
PDBTitle: structure of the pentapeptide repeat protein efsqnr, a dna2 gyrase inhibitor. free amines modified by cyclic3 pentylation with glutaraldehyde.
Phyre2
| 10 |
|
PDB 2j8k chain A domain 1
Region: 139 - 306
Aligned: 168
Modelled: 168
Confidence: 99.9%
Identity: 23%
Fold: Single-stranded right-handed beta-helix
Superfamily: Pentapeptide repeat-like
Family: Pentapeptide repeats
Phyre2
| 11 |
|
PDB 2o6w chain A
Region: 148 - 290
Aligned: 142
Modelled: 143
Confidence: 99.9%
Identity: 14%
PDB header:unknown function
Chain: A: PDB Molecule:repeat five residue (rfr) protein or
PDBTitle: crystal structure of a pentapeptide repeat protein (rfr23)2 from the cyanobacterium cyanothece 51142
Phyre2
| 12 |
|
PDB 3n90 chain A
Region: 145 - 309
Aligned: 140
Modelled: 141
Confidence: 99.9%
Identity: 16%
PDB header:unknown function
Chain: A: PDB Molecule:thylakoid lumenal 15 kda protein 1, chloroplastic;
PDBTitle: the 1.7 angstrom resolution crystal structure of at2g44920, a2 pentapeptide repeat protein from arabidopsis thaliana thylakoid3 lumen.
Phyre2
| 13 |
|
PDB 2g0y chain A
Region: 266 - 395
Aligned: 130
Modelled: 130
Confidence: 99.8%
Identity: 11%
PDB header:unknown function
Chain: A: PDB Molecule:pentapeptide repeat protein;
PDBTitle: crystal structure of a lumenal pentapeptide repeat protein from2 cyanothece sp 51142 at 2.3 angstrom resolution. tetragonal crystal3 form
Phyre2
| 14 |
|
PDB 2qza chain A
Region: 152 - 421
Aligned: 208
Modelled: 217
Confidence: 99.8%
Identity: 14%
PDB header:ligase
Chain: A: PDB Molecule:secreted effector protein;
PDBTitle: crystal structure of salmonella effector protein sopa
Phyre2
| 15 |
|
PDB 2j8i chain B
Region: 261 - 374
Aligned: 114
Modelled: 114
Confidence: 99.8%
Identity: 13%
PDB header:toxin
Chain: B: PDB Molecule:np275;
PDBTitle: structure of np275, a pentapeptide repeat protein from2 nostoc punctiforme
Phyre2
| 16 |
|
PDB 2f3l chain A domain 1
Region: 262 - 389
Aligned: 128
Modelled: 128
Confidence: 99.8%
Identity: 17%
Fold: Single-stranded right-handed beta-helix
Superfamily: Pentapeptide repeat-like
Family: Pentapeptide repeats
Phyre2
| 17 |
|
PDB 2j8i chain A domain 1
Region: 139 - 230
Aligned: 92
Modelled: 92
Confidence: 99.7%
Identity: 18%
Fold: Single-stranded right-handed beta-helix
Superfamily: Pentapeptide repeat-like
Family: Pentapeptide repeats
Phyre2
| 18 |
|
PDB 2y0s chain Q
Region: 62 - 90
Aligned: 29
Modelled: 29
Confidence: 26.4%
Identity: 31%
PDB header:transferase
Chain: Q: PDB Molecule:rna polymerase subunit 13;
PDBTitle: crystal structure of sulfolobus shibatae rna polymerase in2 p21 space group
Phyre2
| 19 |
|
PDB 2y0s chain J
Region: 62 - 90
Aligned: 29
Modelled: 29
Confidence: 26.4%
Identity: 31%
PDB header:transferase
Chain: J: PDB Molecule:rna polymerase subunit 13;
PDBTitle: crystal structure of sulfolobus shibatae rna polymerase in2 p21 space group
Phyre2