1 c2xznQ_
100.0
24
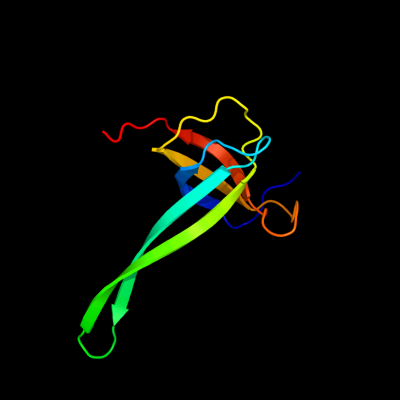
PDB header: ribosomeChain: Q: PDB Molecule: ribosomal protein s17 containing protein;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
2 d2gy9q1
100.0
100
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like3 c3bbnQ_
100.0
37
PDB header: ribosomeChain: Q: PDB Molecule: ribosomal protein s17;PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
4 d2uubq1
100.0
49
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like5 d1i94q_
100.0
49
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like6 c1s1hQ_
100.0
35
PDB header: ribosomeChain: Q: PDB Molecule: 40s ribosomal protein s11;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1h,4 contains 40s subunit. the 60s ribosomal subunit is in file5 1s1i.
7 d1ripa_
100.0
54
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like8 c2zkqq_
100.0
36
PDB header: ribosomal protein/rnaChain: Q: PDB Molecule: PDBTitle: structure of a mammalian ribosomal 40s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
9 c3izbP_
99.8
31
PDB header: ribosomeChain: P: PDB Molecule: 40s ribosomal protein rps11 (s17p);PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
10 c3iz6P_
99.8
24
PDB header: ribosomeChain: P: PDB Molecule: 40s ribosomal protein s11 (s17p);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
11 d1e32a1
26.8
38
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Cdc48 N-terminal domain-like12 c2xzm1_
26.0
50
PDB header: ribosomeChain: 1: PDB Molecule: ribosomal protein s28e containing protein;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
13 c1negA_
23.6
7
PDB header: structural proteinChain: A: PDB Molecule: spectrin alpha chain, brain;PDBTitle: crystal structure analysis of n-and c-terminal labeled sh3-2 domain of alpha-chicken spectrin
14 d1nega_
23.6
7
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain15 d1ue9a_
22.4
14
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain16 d1uuea_
21.7
7
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain17 d1ny4a_
21.6
29
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like18 d1ne3a_
20.5
15
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like19 c2pqhA_
19.9
7
PDB header: structural proteinChain: A: PDB Molecule: spectrin alpha chain, brain;PDBTitle: structure of sh3 chimera with a type ii ligand linked to the chain c-2 terminal
20 c2rqrA_
19.8
25
PDB header: protein bindingChain: A: PDB Molecule: engulfment and cell motility protein 1, linker, dedicatorPDBTitle: the solution structure of human dock2 sh3 domain - elmo1 peptide2 chimera complex
21 d1h8ka_
not modelled
19.8
7
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain22 c2jvvA_
not modelled
19.6
12
PDB header: transcriptionChain: A: PDB Molecule: transcription antitermination protein nusg;PDBTitle: solution structure of e. coli nusg carboxyterminal domain
23 c2kvqG_
not modelled
19.6
12
PDB header: transcriptionChain: G: PDB Molecule: transcription antitermination protein nusg;PDBTitle: solution structure of nuse:nusg-ctd complex
24 d1utia_
not modelled
19.4
17
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain25 d1e6ga_
not modelled
19.3
7
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain26 c2d8jA_
not modelled
19.0
10
PDB header: transferaseChain: A: PDB Molecule: fyn-related kinase;PDBTitle: solution structure of the sh3 domain of fyn-related kinase
27 c3d3rA_
not modelled
18.9
24
PDB header: chaperoneChain: A: PDB Molecule: hydrogenase assembly chaperone hypc/hupf;PDBTitle: crystal structure of the hydrogenase assembly chaperone hypc/hupf2 family protein from shewanella oneidensis mr-1
28 d1nz9a_
not modelled
18.9
21
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: N-utilization substance G protein NusG, C-terminal domain29 c3nmzD_
not modelled
18.6
25
PDB header: cell adhesion/cell cycleChain: D: PDB Molecule: rho guanine nucleotide exchange factor 4;PDBTitle: crytal structure of apc complexed with asef
30 d3d3ra1
not modelled
18.4
24
Fold: OB-foldSuperfamily: HupF/HypC-likeFamily: HupF/HypC-like31 c2dl3A_
not modelled
18.3
16
PDB header: cell adhesion, signaling proteinChain: A: PDB Molecule: sorbin and sh3 domain-containing protein 1;PDBTitle: solution structure of the first sh3 domain of human sorbin2 and sh3 domain-containing protein 1
32 c2dmoA_
not modelled
18.1
10
PDB header: signaling proteinChain: A: PDB Molecule: neutrophil cytosol factor 2;PDBTitle: refined solution structure of the 1st sh3 domain from human2 neutrophil cytosol factor 2 (ncf-2)
33 d2ot2a1
not modelled
17.9
27
Fold: OB-foldSuperfamily: HupF/HypC-likeFamily: HupF/HypC-like34 d1arka_
not modelled
17.6
16
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain35 c2nwmA_
not modelled
17.6
16
PDB header: cell adhesionChain: A: PDB Molecule: vinexin;PDBTitle: solution structure of the first sh3 domain of human vinexin2 and its interaction with the peptides from vinculin
36 d1nppa2
not modelled
17.5
36
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: N-utilization substance G protein NusG, C-terminal domain37 d1gria2
not modelled
17.3
7
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain38 d1k4us_
not modelled
17.3
14
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain39 d1dzfa2
not modelled
17.3
18
Fold: RPB5-like RNA polymerase subunitSuperfamily: RPB5-like RNA polymerase subunitFamily: RPB540 d1u0la1
not modelled
17.1
30
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like41 d1uj0a_
not modelled
17.1
6
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain42 d1udxa3
not modelled
16.9
36
Fold: Obg GTP-binding protein C-terminal domainSuperfamily: Obg GTP-binding protein C-terminal domainFamily: Obg GTP-binding protein C-terminal domain43 c2jteA_
not modelled
16.9
7
PDB header: signaling proteinChain: A: PDB Molecule: cd2-associated protein;PDBTitle: third sh3 domain of cd2ap
44 d1i07a_
not modelled
16.6
21
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain45 c2yupA_
not modelled
16.5
7
PDB header: cell adhesionChain: A: PDB Molecule: vinexin;PDBTitle: solution structure of the second sh3 domain of human vinexin
46 d2z1ca1
not modelled
16.5
22
Fold: OB-foldSuperfamily: HupF/HypC-likeFamily: HupF/HypC-like47 c1zx6A_
not modelled
16.4
14
PDB header: protein bindingChain: A: PDB Molecule: ypr154wp;PDBTitle: high-resolution crystal structure of yeast pin3 sh3 domain
48 c1x2pA_
not modelled
16.0
17
PDB header: transferaseChain: A: PDB Molecule: protein arginine n-methyltransferase 2;PDBTitle: solution structure of the sh3 domain of the protein2 arginine n-methyltransferase 2
49 c3cqtA_
not modelled
15.8
15
PDB header: transferaseChain: A: PDB Molecule: proto-oncogene tyrosine-protein kinase fyn;PDBTitle: n53i v55l mutant of fyn sh3 domain
50 d1hmja_
not modelled
15.7
24
Fold: RPB5-like RNA polymerase subunitSuperfamily: RPB5-like RNA polymerase subunitFamily: RPB551 c2ct3A_
not modelled
15.6
21
PDB header: signaling proteinChain: A: PDB Molecule: vinexin;PDBTitle: solution structure of the sh3 domain of the vinexin protein
52 d1pwta_
not modelled
15.4
6
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain53 c2e5kA_
not modelled
15.3
10
PDB header: protein bindingChain: A: PDB Molecule: suppressor of t-cell receptor signaling 1;PDBTitle: solution structure of sh3 domain in suppressor of t-cell2 receptor signaling 1
54 c2xmfA_
not modelled
15.2
16
PDB header: motor proteinChain: A: PDB Molecule: myosin 1e sh3;PDBTitle: myosin 1e sh3
55 c2crvA_
not modelled
14.7
31
PDB header: translationChain: A: PDB Molecule: translation initiation factor if-2;PDBTitle: solution structure of c-terminal domain of mitochondrial2 translational initiationfactor 2
56 c3p8bB_
not modelled
14.6
25
PDB header: transferase/transcriptionChain: B: PDB Molecule: transcription antitermination protein nusg;PDBTitle: x-ray crystal structure of pyrococcus furiosus transcription2 elongation factor spt4/5
57 d1ppya_
not modelled
14.2
36
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Pyruvoyl dependent aspartate decarboxylase, ADC58 d1e6ha_
not modelled
14.2
7
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain59 c2dl8A_
not modelled
14.1
5
PDB header: signaling proteinChain: A: PDB Molecule: slit-robo rho gtpase-activating protein 2;PDBTitle: solution structure of the sh3 domain of human slit-robo rho2 gtpase-activating protein 2
60 d1t9ha1
not modelled
14.1
13
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like61 c3iz6Y_
not modelled
13.9
21
PDB header: ribosomeChain: Y: PDB Molecule: 40s ribosomal protein s28 (s28e);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
62 c2dlpA_
not modelled
13.8
7
PDB header: structural proteinChain: A: PDB Molecule: kiaa1783 protein;PDBTitle: solution structure of the sh3 domain of human kiaa17832 protein
63 c2d8hA_
not modelled
13.8
10
PDB header: unknown functionChain: A: PDB Molecule: sh3yl1 protein;PDBTitle: solution structure of the sh3 domain of hypothetical2 protein sh3yl1
64 c2c45F_
not modelled
13.6
27
PDB header: lyaseChain: F: PDB Molecule: aspartate 1-decarboxylase precursor;PDBTitle: native precursor of pyruvoyl dependent aspartate2 decarboxylase
65 c3ougA_
not modelled
13.5
45
PDB header: lyaseChain: A: PDB Molecule: aspartate 1-decarboxylase;PDBTitle: crystal structure of cleaved l-aspartate-alpha-decarboxylase from2 francisella tularensis
66 c1pt1B_
not modelled
13.4
36
PDB header: lyaseChain: B: PDB Molecule: aspartate 1-decarboxylase;PDBTitle: unprocessed pyruvoyl dependent aspartate decarboxylase with histidine2 11 mutated to alanine
67 c2krnA_
not modelled
13.3
28
PDB header: signaling proteinChain: A: PDB Molecule: cd2-associated protein;PDBTitle: high resolution structure of the second sh3 domain of cd2ap
68 c2dbkA_
not modelled
13.2
15
PDB header: signaling proteinChain: A: PDB Molecule: crk-like protein;PDBTitle: solution structures of the sh3 domain of human crk-like2 protein
69 c1wxbA_
not modelled
13.1
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: epidermal growth factor receptor pathwayPDBTitle: solution structure of the sh3 domain from human epidermal2 growth factor receptor pathway substrate 8-like protein
70 d1oeba_
not modelled
13.0
18
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain71 d1opka1
not modelled
13.0
10
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain72 d1wpga1
not modelled
12.9
25
Fold: Double-stranded beta-helixSuperfamily: Calcium ATPase, transduction domain AFamily: Calcium ATPase, transduction domain A73 d2uubd1
not modelled
12.9
20
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: Ribosomal protein S474 c3plxB_
not modelled
12.7
36
PDB header: lyaseChain: B: PDB Molecule: aspartate 1-decarboxylase;PDBTitle: the crystal structure of aspartate alpha-decarboxylase from2 campylobacter jejuni subsp. jejuni nctc 11168
75 d2f4la1
not modelled
12.4
18
Fold: CUB-likeSuperfamily: Acetamidase/Formamidase-likeFamily: Acetamidase/Formamidase-like76 c1vc3B_
not modelled
12.4
27
PDB header: lyaseChain: B: PDB Molecule: l-aspartate-alpha-decarboxylase heavy chain;PDBTitle: crystal structure of l-aspartate-alpha-decarboxylase
77 c1uheA_
not modelled
12.4
55
PDB header: lyaseChain: A: PDB Molecule: aspartate 1-decarboxylase alpha chain;PDBTitle: crystal structure of aspartate decarboxylase, isoaspargine complex
78 c2rf0D_
not modelled
12.3
14
PDB header: transferaseChain: D: PDB Molecule: mitogen-activated protein kinase kinase kinase 10;PDBTitle: crystal structure of human mixed lineage kinase map3k10 sh3 domain
79 c2eyxA_
not modelled
12.1
17
PDB header: signaling proteinChain: A: PDB Molecule: v-crk sarcoma virus ct10 oncogene homologPDBTitle: c-terminal sh3 domain of ct10-regulated kinase
80 c2dilA_
not modelled
12.1
10
PDB header: cell adhesionChain: A: PDB Molecule: proline-serine-threonine phosphatase-interactingPDBTitle: solution structure of the sh3 domain of the human proline-2 serine-threonine phosphatase-interacting protein 1
81 d1g31a_
not modelled
12.1
18
Fold: GroES-likeSuperfamily: GroES-likeFamily: GroES82 c2ct4A_
not modelled
11.8
14
PDB header: signaling proteinChain: A: PDB Molecule: cdc42-interacting protein 4;PDBTitle: solution strutcure of the sh3 domain of the cdc42-2 interacting protein 4
83 d1c06a_
not modelled
11.8
20
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: Ribosomal protein S484 c1pyuD_
not modelled
11.7
36
PDB header: lyaseChain: D: PDB Molecule: aspartate 1-decarboxylase alfa chain;PDBTitle: processed aspartate decarboxylase mutant with ser25 mutated to cys
85 c2yuoA_
not modelled
11.7
21
PDB header: signaling proteinChain: A: PDB Molecule: run and tbc1 domain containing 3;PDBTitle: solution structure of the sh3 domain of mouse run and tbc12 domain containing 3
86 c2i0nA_
not modelled
11.6
19
PDB header: structural proteinChain: A: PDB Molecule: class vii unconventional myosin;PDBTitle: structure of dictyostelium discoideum myosin vii sh3 domain2 with adjacent proline rich region
87 d1ov3a1
not modelled
11.5
22
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain88 c1z9zA_
not modelled
11.5
13
PDB header: structural proteinChain: A: PDB Molecule: cytoskeleton assembly control protein sla1;PDBTitle: crystal structure of yeast sla1 sh3 domain 3
89 c2k6dA_
not modelled
11.5
14
PDB header: sh3 domain/ubiquitinChain: A: PDB Molecule: sh3 domain-containing kinase-binding protein 1;PDBTitle: cin85 sh3-c domain in complex with ubiquitin
90 c1yn8E_
not modelled
11.4
21
PDB header: unknown functionChain: E: PDB Molecule: nap1-binding protein 2;PDBTitle: sh3 domain of yeast nbp2
91 d1j3ta_
not modelled
11.3
15
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain92 c2djqA_
not modelled
11.3
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: sh3 domain containing ring finger 2;PDBTitle: the solution structure of the first sh3 domain of mouse sh32 domain containing ring finger 2
93 c2creA_
not modelled
11.2
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hef-like protein;PDBTitle: solution structure of rsgi ruh-036, an sh3 domain from2 human cdna
94 c1w70A_
not modelled
11.1
10
PDB header: sh3 domainChain: A: PDB Molecule: neutrophil cytosol factor 4;PDBTitle: sh3 domain of p40phox complexed with c-terminal polyproline2 region of p47phox
95 c1wi7A_
not modelled
11.0
17
PDB header: protein bindingChain: A: PDB Molecule: sh3-domain kinase binding protein 1;PDBTitle: solution structure of the sh3 domain of sh3-domain kinase2 binding protein 1
96 d2fo0a1
not modelled
11.0
10
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain97 c2cucA_
not modelled
11.0
10
PDB header: signaling proteinChain: A: PDB Molecule: sh3 domain containing ring finger 2;PDBTitle: solution structure of the sh3 domain of the mouse2 hypothetical protein sh3rf2
98 c1zukA_
not modelled
10.9
6
PDB header: contractile proteinChain: A: PDB Molecule: myosin tail region-interacting protein mti1;PDBTitle: yeast bbc1 sh3 domain complexed with a peptide from las17
99 c3mmlD_
not modelled
10.9
24
PDB header: hydrolaseChain: D: PDB Molecule: allophanate hydrolase subunit 1;PDBTitle: allophanate hydrolase complex from mycobacterium smegmatis, msmeg0435-2 msmeg0436