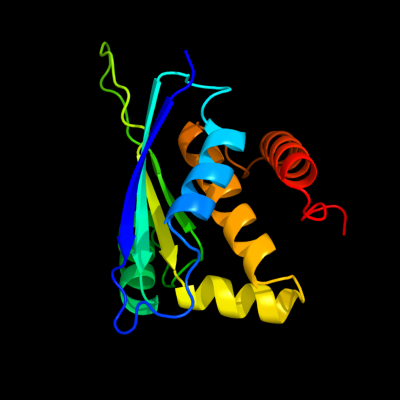
1 d1xm5a_
100.0
100
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Predicted metal-dependent hydrolase2 c1xaxA_
100.0
69
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical upf0054 protein hi0004;PDBTitle: nmr structure of hi0004, a putative essential gene product2 from haemophilus influenzae
3 d1oz9a_
100.0
33
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Predicted metal-dependent hydrolase4 d1tvia_
100.0
28
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Predicted metal-dependent hydrolase5 d1xfia_
65.4
12
Fold: AF1104-likeSuperfamily: AF1104-likeFamily: AF1104-like6 d1lmla_
65.0
19
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Leishmanolysin7 c2xhqA_
59.4
29
PDB header: hydrolaseChain: A: PDB Molecule: archaemetzincin;PDBTitle: crystal structure of archaemetzincin (amza) from archaeoglobus2 fulgidus at 1.45 a resolution
8 c2x7mA_
55.0
19
PDB header: hydrolaseChain: A: PDB Molecule: archaemetzincin;PDBTitle: crystal structure of archaemetzincin (amza) from methanopyrus2 kandleri at 1.5 a resolution
9 d1k7ia2
50.3
11
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Serralysin-like metalloprotease, catalytic (N-terminal) domain10 c1u9pA_
49.2
15
PDB header: unknown functionChain: A: PDB Molecule: parc;PDBTitle: permuted single-chain arc
11 c1qysA_
45.2
33
PDB header: de novo proteinChain: A: PDB Molecule: top7;PDBTitle: crystal structure of top7: a computationally designed2 protein with a novel fold
12 d1wgna_
44.5
15
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain13 c2jvfA_
44.4
33
PDB header: de novo proteinChain: A: PDB Molecule: de novo protein m7;PDBTitle: solution structure of m7, a computationally-designed2 artificial protein
14 d1asta_
41.1
25
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Astacin15 c3edhA_
41.0
35
PDB header: hydrolaseChain: A: PDB Molecule: bone morphogenetic protein 1;PDBTitle: crystal structure of bone morphogenetic protein 1 protease2 domain in complex with partially bound dmso
16 c3lqbA_
40.5
29
PDB header: hydrolaseChain: A: PDB Molecule: loc792177 protein;PDBTitle: crystal structure of the hatching enzyme zhe1 from the zebrafish danio2 rerio
17 d1hv5a_
40.4
25
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Matrix metalloproteases, catalytic domain18 d1y93a1
40.3
35
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Matrix metalloproteases, catalytic domain19 d1fbla2
38.8
30
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Matrix metalloproteases, catalytic domain20 d2ovxa1
38.2
30
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Matrix metalloproteases, catalytic domain21 d1cgla_
not modelled
36.8
30
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Matrix metalloproteases, catalytic domain22 d1hova_
not modelled
36.2
25
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Matrix metalloproteases, catalytic domain23 d1hfca_
not modelled
36.1
30
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Matrix metalloproteases, catalytic domain24 d1xuca1
not modelled
34.9
35
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Matrix metalloproteases, catalytic domain25 d1q3aa_
not modelled
34.8
25
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Matrix metalloproteases, catalytic domain26 c2jsdA_
not modelled
34.1
30
PDB header: hydrolaseChain: A: PDB Molecule: matrix metalloproteinase-20;PDBTitle: solution structure of mmp20 complexed with nngh
27 d1rm8a_
not modelled
32.5
35
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Matrix metalloproteases, catalytic domain28 d1i76a_
not modelled
31.1
30
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Matrix metalloproteases, catalytic domain29 d1qiba_
not modelled
30.6
25
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Matrix metalloproteases, catalytic domain30 c3lq0A_
not modelled
28.5
27
PDB header: hydrolaseChain: A: PDB Molecule: proastacin;PDBTitle: zymogen structure of crayfish astacin metallopeptidase
31 c3lmcA_
not modelled
28.3
29
PDB header: hydrolaseChain: A: PDB Molecule: peptidase, zinc-dependent;PDBTitle: crystal structure of zinc-dependent peptidase from methanocorpusculum2 labreanum (strain z), northeast structural genomics consortium target3 mur16
32 c2xs4A_
not modelled
27.2
30
PDB header: hydrolaseChain: A: PDB Molecule: karilysin protease;PDBTitle: structure of karilysin catalytic mmp domain in complex with2 magnesium
33 d1mmqa_
not modelled
25.9
30
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Matrix metalloproteases, catalytic domain34 c1slmA_
not modelled
25.3
25
PDB header: hydrolaseChain: A: PDB Molecule: stromelysin-1;PDBTitle: crystal structure of fibroblast stromelysin-1: the c-truncated human2 proenzyme
35 c3qyyB_
not modelled
24.6
12
PDB header: signaling protein/inhibitorChain: B: PDB Molecule: response regulator;PDBTitle: a novel interaction mode between a microbial ggdef domain and the bis-2 (3, 5 )-cyclic di-gmp
36 d1cxva_
not modelled
24.4
35
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Matrix metalloproteases, catalytic domain37 c3i5bA_
not modelled
21.5
24
PDB header: signaling proteinChain: A: PDB Molecule: wspr response regulator;PDBTitle: crystal structure of the isolated ggdef domain of wpsr from2 pseudomonas aeruginosa
38 d1hy7a_
not modelled
20.9
25
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Matrix metalloproteases, catalytic domain39 d1eaka2
not modelled
18.8
25
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Matrix metalloproteases, catalytic domain40 c3hvaA_
not modelled
18.6
21
PDB header: transferaseChain: A: PDB Molecule: protein fimx;PDBTitle: crystal structure of fimx ggdef domain from pseudomonas2 aeruginosa
41 c2a97B_
not modelled
17.0
17
PDB header: hydrolaseChain: B: PDB Molecule: botulinum neurotoxin type f;PDBTitle: crystal structure of catalytic domain of clostridium2 botulinum neurotoxin serotype f
42 d1w25a3
not modelled
15.6
17
Fold: Ferredoxin-likeSuperfamily: Nucleotide cyclaseFamily: GGDEF domain43 c1jmtB_
not modelled
13.2
31
PDB header: rna binding proteinChain: B: PDB Molecule: splicing factor u2af 65 kda subunit;PDBTitle: x-ray structure of a core u2af65/u2af35 heterodimer
44 c3i5cA_
not modelled
11.2
24
PDB header: signaling proteinChain: A: PDB Molecule: fusion of general control protein gcn4 and wspr responsePDBTitle: crystal structure of a fusion protein containing the leucine zipper of2 gcn4 and the ggdef domain of wspr from pseudomonas aeruginosa
45 c1om8A_
not modelled
10.9
12
PDB header: hydrolaseChain: A: PDB Molecule: serralysin;PDBTitle: crystal structure of a cold adapted alkaline protease from pseudomonas2 tac ii 18, co-crystallyzed with 10 mm edta
46 c1dowB_
not modelled
10.1
14
PDB header: cell adhesionChain: B: PDB Molecule: beta-catenin;PDBTitle: crystal structure of a chimera of beta-catenin and alpha-2 catenin
47 c3ba0A_
not modelled
10.0
35
PDB header: hydrolaseChain: A: PDB Molecule: macrophage metalloelastase;PDBTitle: crystal structure of full-length human mmp-12
48 c3bkdC_
not modelled
9.7
50
PDB header: viral protein, membrane proteinChain: C: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
49 c3bkdH_
not modelled
9.7
50
PDB header: viral protein, membrane proteinChain: H: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
50 c3bkdB_
not modelled
9.7
50
PDB header: viral protein, membrane proteinChain: B: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
51 c3bkdA_
not modelled
9.7
50
PDB header: viral protein, membrane proteinChain: A: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
52 c3bkdG_
not modelled
9.7
50
PDB header: viral protein, membrane proteinChain: G: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
53 c3bkdD_
not modelled
9.7
50
PDB header: viral protein, membrane proteinChain: D: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
54 c3bkdE_
not modelled
9.7
50
PDB header: viral protein, membrane proteinChain: E: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
55 c3bkdF_
not modelled
9.7
50
PDB header: viral protein, membrane proteinChain: F: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
56 d1sata2
not modelled
9.7
18
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Serralysin-like metalloprotease, catalytic (N-terminal) domain57 d3e11a1
not modelled
9.6
30
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: TTHA0227-like58 d1gtda_
not modelled
9.1
30
Fold: PurS-likeSuperfamily: PurS-likeFamily: PurS subunit of FGAM synthetase59 d1bqqm_
not modelled
8.8
35
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Matrix metalloproteases, catalytic domain60 c2rjqA_
not modelled
8.7
38
PDB header: hydrolaseChain: A: PDB Molecule: adamts-5;PDBTitle: crystal structure of adamts5 with inhibitor bound
61 c3iclA_
not modelled
8.5
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: eal/ggdef domain protein;PDBTitle: x-ray structure of protein (eal/ggdef domain protein) from2 m.capsulatus, northeast structural genomics consortium3 target mcr174c
62 c2erpA_
not modelled
8.5
38
PDB header: toxinChain: A: PDB Molecule: vascular apoptosis-inducing protein 1;PDBTitle: crystal structure of vascular apoptosis-inducing protein-1(inhibitor-2 bound form)
63 d1kufa_
not modelled
8.4
30
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Reprolysin-like64 c3ignA_
not modelled
8.1
19
PDB header: transferaseChain: A: PDB Molecule: diguanylate cyclase;PDBTitle: crystal structure of the ggdef domain from marinobacter2 aquaeolei diguanylate cyclase complexed with c-di-gmp -3 northeast structural genomics consortium target mqr89a
65 c2gjhA_
not modelled
7.7
35
PDB header: de novo proteinChain: A: PDB Molecule: designed protein;PDBTitle: nmr structure of cfr (c-terminal fragment of2 computationally designed novel-topology protein top7)
66 c3gr1A_
not modelled
7.6
10
PDB header: membrane proteinChain: A: PDB Molecule: protein prgh;PDBTitle: periplamic domain of the t3ss inner membrane protein prgh2 from s.typhimurium (fragment 170-392)
67 c2cltB_
not modelled
7.5
30
PDB header: hydrolaseChain: B: PDB Molecule: interstitial collagenase;PDBTitle: crystal structure of the active form (full-length) of human2 fibroblast collagenase.
68 c2zw2B_
not modelled
7.5
14
PDB header: ligaseChain: B: PDB Molecule: putative uncharacterized protein sts178;PDBTitle: crystal structure of formylglycinamide ribonucleotide amidotransferase2 iii from sulfolobus tokodaii (stpurs)
69 d1atla_
not modelled
7.5
25
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Reprolysin-like70 c2dgbA_
not modelled
7.4
42
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein purs;PDBTitle: structure of thermus thermophilus purs in the p21 form
71 d1k1fa_
not modelled
7.4
16
Fold: Bcr-Abl oncoprotein oligomerization domainSuperfamily: Bcr-Abl oncoprotein oligomerization domainFamily: Bcr-Abl oncoprotein oligomerization domain72 c3pjwA_
not modelled
7.3
21
PDB header: lyaseChain: A: PDB Molecule: cyclic dimeric gmp binding protein;PDBTitle: structure of pseudomonas fluorescence lapd ggdef-eal dual domain, i23
73 c3b8zB_
not modelled
7.0
38
PDB header: hydrolaseChain: B: PDB Molecule: protein adamts-5;PDBTitle: high resolution crystal structure of the catalytic domain2 of adamts-5 (aggrecanase-2)
74 c2dw1B_
not modelled
6.8
30
PDB header: apoptosis, toxinChain: B: PDB Molecule: catrocollastatin;PDBTitle: crystal structure of vap2 from crotalus atrox venom (form 2-2 crystal)
75 c3e5aB_
not modelled
6.7
17
PDB header: transferaseChain: B: PDB Molecule: targeting protein for xklp2;PDBTitle: crystal structure of aurora a in complex with vx-680 and tpx2
76 d1r55a_
not modelled
6.7
33
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Reprolysin-like77 d1t4aa_
not modelled
6.7
31
Fold: PurS-likeSuperfamily: PurS-likeFamily: PurS subunit of FGAM synthetase78 c3c0tB_
not modelled
6.4
67
PDB header: transcriptionChain: B: PDB Molecule: mediator of rna polymerase ii transcriptionPDBTitle: structure of the schizosaccharomyces pombe mediator2 subcomplex med8c/18
79 c1p9iA_
not modelled
6.4
21
PDB header: unknown functionChain: A: PDB Molecule: cortexillin i/gcn4 hybrid peptide;PDBTitle: coiled-coil x-ray structure at 1.17 a resolution
80 d2b7ea1
not modelled
6.2
42
Fold: Another 3-helical bundleSuperfamily: FF domainFamily: FF domain81 c3ipfA_
not modelled
6.2
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the q251q8_deshy protein from desulfitobacterium2 hafniense. northeast structural genomics consortium target dhr8c.
82 c2awyB_
not modelled
6.1
13
PDB header: oxygen storage/transportChain: B: PDB Molecule: hemerythrin-like domain protein dcrh;PDBTitle: met-dcrh-hr
83 d2a6aa2
not modelled
6.1
29
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: YeaZ-like84 d1qf6a4
not modelled
6.0
19
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain85 c3o7kA_
not modelled
6.0
7
PDB header: lyaseChain: A: PDB Molecule: ohcu decarboxylase;PDBTitle: crystal structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline2 decarboxylase from klebsiella pneumoniae
86 c3b6nA_
not modelled
5.8
14
PDB header: lyaseChain: A: PDB Molecule: 2-c-methyl-d-erythritol 2,4-cyclodiphosphatePDBTitle: crystal structure of 2c-methyl-d-erythritol 2,4-2 cyclodiphosphate synthase pv003920 from plasmodium vivax
87 c2ppwA_
not modelled
5.8
11
PDB header: isomeraseChain: A: PDB Molecule: conserved domain protein;PDBTitle: the crystal structure of uncharacterized ribose 5-phosphate isomerase2 rpib from streptococcus pneumoniae
88 c3f6nA_
not modelled
5.8
29
PDB header: viral protein, dna-binding proteinChain: A: PDB Molecule: virion-associated protein;PDBTitle: crystal structure of the virion-associated protein p3 from2 caulimovirus
89 d1kapp2
not modelled
5.6
30
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Serralysin-like metalloprotease, catalytic (N-terminal) domain90 d2g8la1
not modelled
5.6
13
Fold: AF1104-likeSuperfamily: AF1104-likeFamily: AF1104-like91 c2kadA_
not modelled
5.6
40
PDB header: membrane proteinChain: A: PDB Molecule: transmembrane peptide of matrix protein 2;PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain
92 c2kadB_
not modelled
5.6
40
PDB header: membrane proteinChain: B: PDB Molecule: transmembrane peptide of matrix protein 2;PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain
93 c2kadD_
not modelled
5.6
40
PDB header: membrane proteinChain: D: PDB Molecule: transmembrane peptide of matrix protein 2;PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain
94 c2kadC_
not modelled
5.6
40
PDB header: membrane proteinChain: C: PDB Molecule: transmembrane peptide of matrix protein 2;PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain
95 c2v4bB_
not modelled
5.6
18
PDB header: hydrolaseChain: B: PDB Molecule: adamts-1;PDBTitle: crystal structure of human adamts-1 catalytic domain and2 cysteine-rich domain (apo-form)
96 c3dl1A_
not modelled
5.5
27
PDB header: hydrolaseChain: A: PDB Molecule: putative metal-dependent hydrolase;PDBTitle: crystal structure of a putative metal-dependent hydrolase2 (yp_001336084.1) from klebsiella pneumoniae subsp. pneumoniae mgh3 78578 at 2.20 a resolution
97 c1fdfA_
not modelled
5.5
29
PDB header: signaling proteinChain: A: PDB Molecule: rhodopsin;PDBTitle: helix 7 bovine rhodopsin
98 d1qmya_
not modelled
5.4
38
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: FMDV leader protease99 c3k4tB_
not modelled
5.4
29
PDB header: viral protein, dna-binding proteinChain: B: PDB Molecule: virion-associated protein;PDBTitle: crystal structure of the virion-associated protein p3 from2 caulimovirus