| 1 |
|
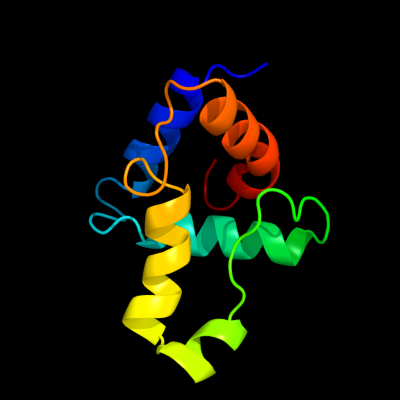
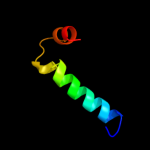
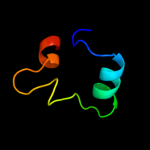
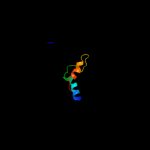
PDB 3c4r chain C
Region: 2 - 95
Aligned: 84
Modelled: 94
Confidence: 100.0%
Identity: 36%
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of an uncharacterized protein encoded by2 cryptic prophage
Phyre2
| 2 |
|
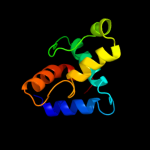
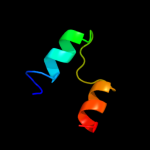
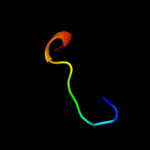
PDB 2qxv chain B
Region: 63 - 70
Aligned: 8
Modelled: 8
Confidence: 21.3%
Identity: 75%
PDB header:gene regulation
Chain: B: PDB Molecule:enhancer of zeste homolog 2;
PDBTitle: structural basis of ezh2 recognition by eed
Phyre2
| 3 |
|
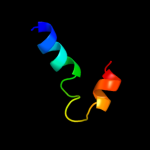
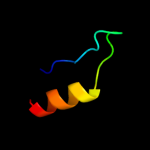
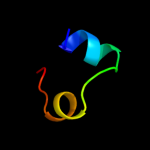
PDB 1y7q chain A domain 1
Region: 62 - 87
Aligned: 25
Modelled: 26
Confidence: 19.9%
Identity: 12%
Fold: Acyl carrier protein-like
Superfamily: Retrovirus capsid dimerization domain-like
Family: SCAN domain
Phyre2
| 4 |
|
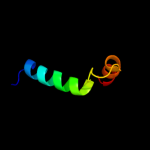
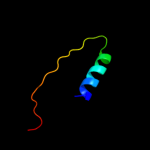
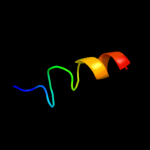
PDB 2fi2 chain A domain 1
Region: 54 - 87
Aligned: 33
Modelled: 34
Confidence: 19.7%
Identity: 15%
Fold: Acyl carrier protein-like
Superfamily: Retrovirus capsid dimerization domain-like
Family: SCAN domain
Phyre2
| 5 |
|
PDB 3lhr chain A
Region: 54 - 87
Aligned: 33
Modelled: 34
Confidence: 15.7%
Identity: 18%
PDB header:transcription regulator
Chain: A: PDB Molecule:zinc finger protein 24;
PDBTitle: crystal structure of the scan domain from human znf24
Phyre2
| 6 |
|
PDB 3l6a chain A
Region: 63 - 93
Aligned: 30
Modelled: 31
Confidence: 15.6%
Identity: 17%
PDB header:translation
Chain: A: PDB Molecule:eukaryotic translation initiation factor 4 gamma 2;
PDBTitle: crystal structure of the c-terminal region of human p97
Phyre2
| 7 |
|
PDB 3e96 chain B
Region: 43 - 67
Aligned: 25
Modelled: 25
Confidence: 13.9%
Identity: 20%
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 bacillus clausii
Phyre2
| 8 |
|
PDB 2ek0 chain B
Region: 21 - 53
Aligned: 33
Modelled: 33
Confidence: 12.5%
Identity: 12%
PDB header:metal binding protein
Chain: B: PDB Molecule:stage v sporulation protein s (spovs) related protein;
PDBTitle: stage v sporolation protein s (spovs) from thermus thermophilus zinc2 form
Phyre2
| 9 |
|
PDB 2l53 chain B
Region: 19 - 34
Aligned: 15
Modelled: 16
Confidence: 11.5%
Identity: 40%
PDB header:ca-binding protein/proton transport
Chain: B: PDB Molecule:voltage-gated sodium channel type v alpha isoform b
PDBTitle: solution nmr structure of apo-calmodulin in complex with the iq motif2 of human cardiac sodium channel nav1.5
Phyre2
| 10 |
|
PDB 3bd1 chain B
Region: 58 - 96
Aligned: 33
Modelled: 39
Confidence: 10.3%
Identity: 27%
PDB header:transcription
Chain: B: PDB Molecule:cro protein;
PDBTitle: structure of the cro protein from putative prophage element xfaso 1 in2 xylella fastidiosa strain ann-1
Phyre2
| 11 |
|
PDB 1pu1 chain A
Region: 21 - 37
Aligned: 17
Modelled: 17
Confidence: 9.4%
Identity: 12%
Fold: Hypothetical protein MTH677
Superfamily: Hypothetical protein MTH677
Family: Hypothetical protein MTH677
Phyre2
| 12 |
|
PDB 3fgg chain A
Region: 50 - 60
Aligned: 11
Modelled: 11
Confidence: 8.7%
Identity: 45%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein bce2196;
PDBTitle: crystal structure of putative ecf-type sigma factor negative effector2 from bacillus cereus
Phyre2
| 13 |
|
PDB 2a2j chain A domain 1
Region: 49 - 73
Aligned: 25
Modelled: 25
Confidence: 8.5%
Identity: 24%
Fold: Split barrel-like
Superfamily: FMN-binding split barrel
Family: PNP-oxidase like
Phyre2
| 14 |
|
PDB 1tp6 chain A
Region: 66 - 87
Aligned: 20
Modelled: 22
Confidence: 8.1%
Identity: 30%
Fold: Cystatin-like
Superfamily: NTF2-like
Family: PA1314-like
Phyre2
| 15 |
|
PDB 2kxw chain B
Region: 18 - 34
Aligned: 15
Modelled: 17
Confidence: 8.0%
Identity: 33%
PDB header:calcium-binding protein/metal transport
Chain: B: PDB Molecule:sodium channel protein type 2 subunit alpha;
PDBTitle: structure of the c-domain fragment of apo calmodulin bound to the iq2 motif of nav1.2
Phyre2
| 16 |
|
PDB 1xdt chain T
Region: 54 - 96
Aligned: 35
Modelled: 43
Confidence: 7.4%
Identity: 26%
PDB header:complex (toxin/growth factor)
Chain: T: PDB Molecule:diphtheria toxin;
PDBTitle: complex of diphtheria toxin and heparin-binding epidermal growth2 factor
Phyre2
| 17 |
|
PDB 1ijw chain C
Region: 2 - 30
Aligned: 27
Modelled: 29
Confidence: 7.0%
Identity: 15%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: Recombinase DNA-binding domain
Phyre2
| 18 |
|
PDB 2nr5 chain A domain 1
Region: 21 - 77
Aligned: 55
Modelled: 0
Confidence: 7.0%
Identity: 20%
Fold: Ferritin-like
Superfamily: SO2669-like
Family: SO2669-like
Phyre2
| 19 |
|
PDB 1ug3 chain A domain 2
Region: 61 - 94
Aligned: 33
Modelled: 34
Confidence: 6.7%
Identity: 24%
Fold: alpha-alpha superhelix
Superfamily: ARM repeat
Family: MIF4G domain-like
Phyre2
| 20 |
|
PDB 2ow7 chain A
Region: 65 - 87
Aligned: 23
Modelled: 23
Confidence: 6.7%
Identity: 17%
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-mannosidase 2;
PDBTitle: golgi alpha-mannosidase ii complex with (1r,6s,7r,8s)-1-2 thioniabicyclo[4.3.0]nonan-7,8-diol chloride
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|