| 1 |
|
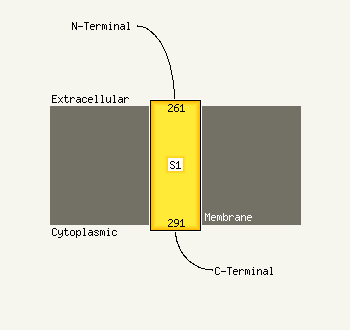
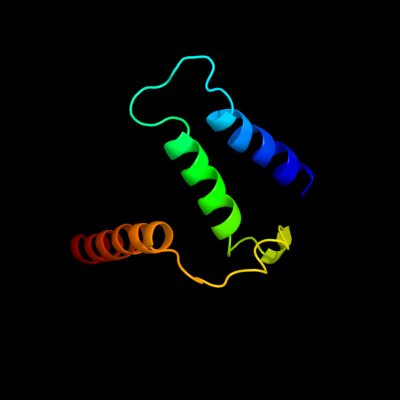
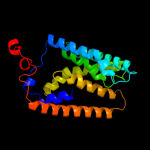
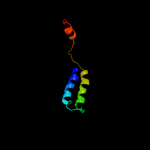
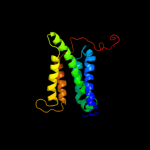
PDB 1s7b chain A
Region: 224 - 302
Aligned: 71
Modelled: 79
Confidence: 97.5%
Identity: 15%
Fold: Multidrug resistance efflux transporter EmrE
Superfamily: Multidrug resistance efflux transporter EmrE
Family: Multidrug resistance efflux transporter EmrE
Phyre2
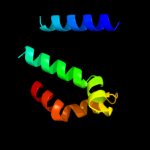
| 2 |
|
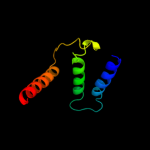
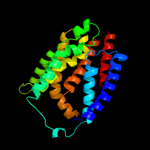
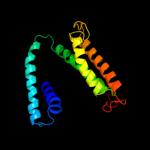
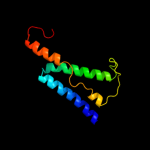
PDB 2i68 chain B
Region: 228 - 299
Aligned: 58
Modelled: 72
Confidence: 75.0%
Identity: 17%
PDB header:transport protein
Chain: B: PDB Molecule:protein emre;
PDBTitle: cryo-em based theoretical model structure of transmembrane2 domain of the multidrug-resistance antiporter from e. coli3 emre
Phyre2
| 3 |
|
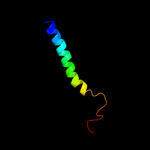
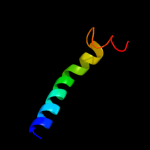
PDB 3b9y chain A
Region: 44 - 321
Aligned: 221
Modelled: 221
Confidence: 63.1%
Identity: 13%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
Phyre2
| 4 |
|
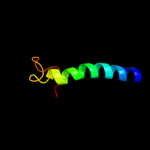
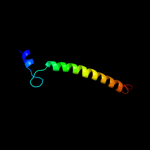
PDB 3hd6 chain A
Region: 2 - 231
Aligned: 230
Modelled: 230
Confidence: 25.9%
Identity: 13%
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
Phyre2
| 5 |
|
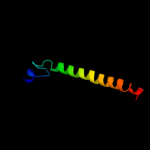
PDB 2knc chain A
Region: 281 - 319
Aligned: 39
Modelled: 39
Confidence: 21.0%
Identity: 13%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 6 |
|
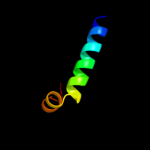
PDB 2jo1 chain A
Region: 284 - 321
Aligned: 38
Modelled: 38
Confidence: 20.4%
Identity: 8%
PDB header:hydrolase regulator
Chain: A: PDB Molecule:phospholemman;
PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
Phyre2
| 7 |
|
PDB 2akh chain Z
Region: 260 - 321
Aligned: 62
Modelled: 62
Confidence: 17.2%
Identity: 19%
PDB header:protein transport
Chain: Z: PDB Molecule:preprotein translocase sece subunit;
PDBTitle: normal mode-based flexible fitted coordinates of a non-2 translocating secyeg protein-conducting channel into the3 cryo-em map of a secyeg-nascent chain-70s ribosome complex4 from e. coli
Phyre2
| 8 |
|
PDB 3dh4 chain A
Region: 160 - 321
Aligned: 154
Modelled: 162
Confidence: 15.1%
Identity: 7%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
Phyre2
| 9 |
|
PDB 2jp3 chain A
Region: 284 - 317
Aligned: 34
Modelled: 34
Confidence: 14.4%
Identity: 12%
PDB header:transcription
Chain: A: PDB Molecule:fxyd domain-containing ion transport regulator 4;
PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
Phyre2
| 10 |
|
PDB 1zrt chain D
Region: 258 - 312
Aligned: 50
Modelled: 55
Confidence: 12.6%
Identity: 12%
PDB header:oxidoreductase/metal transport
Chain: D: PDB Molecule:cytochrome c1;
PDBTitle: rhodobacter capsulatus cytochrome bc1 complex with2 stigmatellin bound
Phyre2
| 11 |
|
PDB 3cwb chain Q
Region: 258 - 318
Aligned: 56
Modelled: 61
Confidence: 8.1%
Identity: 9%
PDB header:oxidoreductase
Chain: Q: PDB Molecule:mitochondrial cytochrome c1, heme protein;
PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
Phyre2
| 12 |
|
PDB 1p84 chain D
Region: 258 - 311
Aligned: 49
Modelled: 54
Confidence: 7.8%
Identity: 10%
PDB header:oxidoreductase
Chain: D: PDB Molecule:cytochrome c1, heme protein;
PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
Phyre2
| 13 |
|
PDB 2rdd chain B
Region: 280 - 316
Aligned: 37
Modelled: 37
Confidence: 7.2%
Identity: 14%
PDB header:membrane protein/transport protein
Chain: B: PDB Molecule:upf0092 membrane protein yajc;
PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
Phyre2
| 14 |
|
PDB 1iwg chain A domain 7
Region: 221 - 321
Aligned: 93
Modelled: 101
Confidence: 6.7%
Identity: 11%
Fold: Multidrug efflux transporter AcrB transmembrane domain
Superfamily: Multidrug efflux transporter AcrB transmembrane domain
Family: Multidrug efflux transporter AcrB transmembrane domain
Phyre2
| 15 |
|
PDB 2gfp chain A
Region: 151 - 321
Aligned: 166
Modelled: 171
Confidence: 6.6%
Identity: 7%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 16 |
|
PDB 3aqp chain B
Region: 216 - 314
Aligned: 91
Modelled: 99
Confidence: 6.5%
Identity: 10%
PDB header:membrane protein
Chain: B: PDB Molecule:probable secdf protein-export membrane protein;
PDBTitle: crystal structure of secdf, a translocon-associated membrane protein,2 from thermus thrmophilus
Phyre2
| 17 |
|
PDB 2fyn chain H
Region: 258 - 308
Aligned: 46
Modelled: 51
Confidence: 6.2%
Identity: 15%
PDB header:oxidoreductase
Chain: H: PDB Molecule:cytochrome c1;
PDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
Phyre2
| 18 |
|
PDB 1y5i chain C domain 1
Region: 285 - 315
Aligned: 31
Modelled: 31
Confidence: 6.1%
Identity: 13%
Fold: Heme-binding four-helical bundle
Superfamily: Respiratory nitrate reductase 1 gamma chain
Family: Respiratory nitrate reductase 1 gamma chain
Phyre2
| 19 |
|
PDB 2yiu chain E
Region: 258 - 308
Aligned: 46
Modelled: 51
Confidence: 5.6%
Identity: 11%
PDB header:oxidoreductase
Chain: E: PDB Molecule:cytochrome c1, heme protein;
PDBTitle: x-ray structure of the dimeric cytochrome bc1 complex from2 the soil bacterium paracoccus denitrificans at 2.73 angstrom resolution
Phyre2
| 20 |
|
PDB 2b2h chain A
Region: 8 - 244
Aligned: 231
Modelled: 228
Confidence: 5.2%
Identity: 13%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter;
PDBTitle: ammonium transporter amt-1 from a. fulgidus (as)
Phyre2
| 21 |
|
| 22 |
|