| 1 |
|
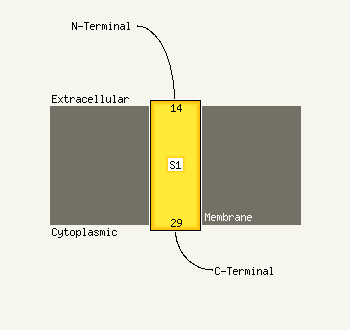
PDB 3jwd chain A
Region: 61 - 79
Aligned: 19
Modelled: 19
Confidence: 18.5%
Identity: 63%
PDB header:viral protein
Chain: A: PDB Molecule:hiv-1 gp120 envelope glycoprotein;
PDBTitle: structure of hiv-1 gp120 with gp41-interactive region: layered2 architecture and basis of conformational mobility
Phyre2
| 2 |
|
PDB 3ngb chain I
Region: 61 - 79
Aligned: 19
Modelled: 19
Confidence: 15.5%
Identity: 58%
PDB header:viral protein/immune system
Chain: I: PDB Molecule:envelope glycoprotein gp160;
PDBTitle: crystal structure of broadly and potently neutralizing antibody vrc012 in complex with hiv-1 gp120
Phyre2
| 3 |
|
PDB 1oe1 chain A domain 2
Region: 4 - 46
Aligned: 40
Modelled: 43
Confidence: 14.9%
Identity: 18%
Fold: Cupredoxin-like
Superfamily: Cupredoxins
Family: Multidomain cupredoxins
Phyre2
| 4 |
|
PDB 4a02 chain A
Region: 9 - 51
Aligned: 43
Modelled: 43
Confidence: 11.6%
Identity: 19%
PDB header:chitin binding protein
Chain: A: PDB Molecule:chitin binding protein;
PDBTitle: x-ray crystallographic structure of efcbm33a
Phyre2
| 5 |
|
PDB 2bf1 chain A
Region: 35 - 44
Aligned: 10
Modelled: 10
Confidence: 10.9%
Identity: 30%
PDB header:virus protein
Chain: A: PDB Molecule:exterior membrane glycoprotein gp120;
PDBTitle: structure of an unliganded and fully-glycosylated siv gp1202 envelope glycoprotein
Phyre2
| 6 |
|
PDB 1tvc chain A domain 1
Region: 7 - 75
Aligned: 64
Modelled: 69
Confidence: 10.3%
Identity: 20%
Fold: Reductase/isomerase/elongation factor common domain
Superfamily: Riboflavin synthase domain-like
Family: Ferredoxin reductase FAD-binding domain-like
Phyre2
| 7 |
|
PDB 2b4c chain G domain 1
Region: 4 - 44
Aligned: 27
Modelled: 27
Confidence: 8.1%
Identity: 41%
Fold: gp120 core
Superfamily: gp120 core
Family: gp120 core
Phyre2
| 8 |
|
PDB 2fgy chain A
Region: 16 - 38
Aligned: 20
Modelled: 23
Confidence: 6.7%
Identity: 55%
PDB header:lyase
Chain: A: PDB Molecule:carboxysome shell polypeptide;
PDBTitle: beta carbonic anhydrase from the carboxysomal shell of2 halothiobacillus neapolitanus (csosca)
Phyre2
| 9 |
|
PDB 1gvh chain A domain 2
Region: 13 - 79
Aligned: 63
Modelled: 67
Confidence: 6.3%
Identity: 14%
Fold: Reductase/isomerase/elongation factor common domain
Superfamily: Riboflavin synthase domain-like
Family: Ferredoxin reductase FAD-binding domain-like
Phyre2
| 10 |
|
PDB 1fqj chain C
Region: 67 - 79
Aligned: 13
Modelled: 13
Confidence: 6.3%
Identity: 54%
PDB header:signaling protein
Chain: C: PDB Molecule:retinal rod rhodopsin-sensitive cgmp 3',5'-
PDBTitle: crystal structure of the heterotrimeric complex of the rgs2 domain of rgs9, the gamma subunit of phosphodiesterase and3 the gt/i1 chimera alpha subunit [(rgs9)-(pdegamma)-4 (gt/i1alpha)-(gdp)-(alf4-)-(mg2+)]
Phyre2
| 11 |
|
PDB 1w07 chain A domain 3
Region: 54 - 112
Aligned: 52
Modelled: 59
Confidence: 5.4%
Identity: 23%
Fold: Acyl-CoA dehydrogenase NM domain-like
Superfamily: Acyl-CoA dehydrogenase NM domain-like
Family: acyl-CoA oxidase N-terminal domains
Phyre2
| 12 |
|
PDB 2bem chain A
Region: 9 - 51
Aligned: 43
Modelled: 43
Confidence: 5.0%
Identity: 21%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: E set domains
Family: E-set domains of sugar-utilizing enzymes
Phyre2