| 1 |
|
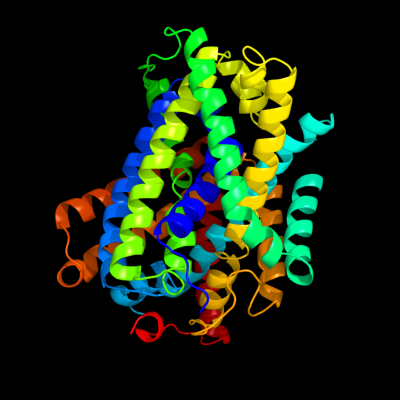
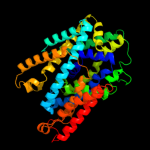
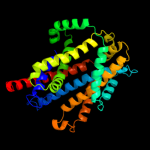
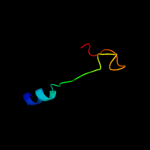
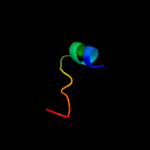
PDB 3gia chain A
Region: 15 - 452
Aligned: 424
Modelled: 424
Confidence: 100.0%
Identity: 20%
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized protein mj0609;
PDBTitle: crystal structure of apct transporter
Phyre2
| 2 |
|
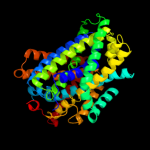
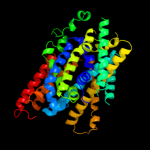
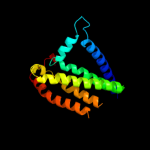
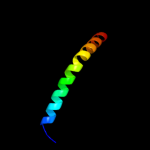
PDB 3lrc chain C
Region: 15 - 445
Aligned: 397
Modelled: 397
Confidence: 100.0%
Identity: 16%
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
Phyre2
| 3 |
|
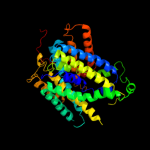
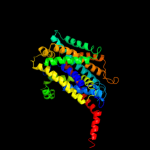
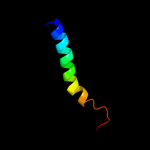
PDB 2jln chain A
Region: 2 - 450
Aligned: 439
Modelled: 439
Confidence: 100.0%
Identity: 11%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
| 4 |
|
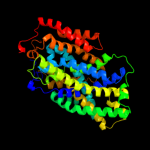
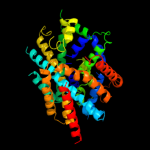
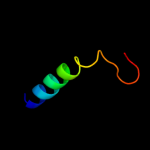
PDB 2xq2 chain A
Region: 16 - 451
Aligned: 419
Modelled: 436
Confidence: 99.5%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
Phyre2
| 5 |
|
PDB 3dh4 chain A
Region: 16 - 450
Aligned: 425
Modelled: 425
Confidence: 99.5%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
Phyre2
| 6 |
|
PDB 2w8a chain C
Region: 5 - 404
Aligned: 389
Modelled: 396
Confidence: 97.1%
Identity: 13%
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
Phyre2
| 7 |
|
PDB 2a65 chain A domain 1
Region: 19 - 442
Aligned: 417
Modelled: 417
Confidence: 96.3%
Identity: 10%
Fold: SNF-like
Superfamily: SNF-like
Family: SNF-like
Phyre2
| 8 |
|
PDB 3hfx chain A
Region: 52 - 390
Aligned: 333
Modelled: 339
Confidence: 92.3%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:l-carnitine/gamma-butyrobetaine antiporter;
PDBTitle: crystal structure of carnitine transporter
Phyre2
| 9 |
|
PDB 3m7b chain A
Region: 283 - 450
Aligned: 160
Modelled: 168
Confidence: 33.5%
Identity: 11%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:tellurite resistance protein teha homolog;
PDBTitle: crystal structure of plant slac1 homolog teha
Phyre2
| 10 |
|
PDB 2rdd chain B
Region: 424 - 451
Aligned: 28
Modelled: 28
Confidence: 23.6%
Identity: 14%
PDB header:membrane protein/transport protein
Chain: B: PDB Molecule:upf0092 membrane protein yajc;
PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
Phyre2
| 11 |
|
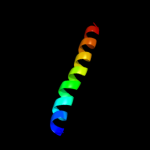
PDB 1ujl chain A
Region: 8 - 29
Aligned: 22
Modelled: 22
Confidence: 20.7%
Identity: 5%
PDB header:membrane protein
Chain: A: PDB Molecule:potassium voltage-gated channel subfamily h
PDBTitle: solution structure of the herg k+ channel s5-p2 extracellular linker
Phyre2
| 12 |
|
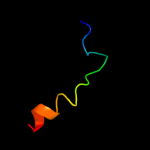
PDB 2jo1 chain A
Region: 420 - 450
Aligned: 31
Modelled: 31
Confidence: 12.1%
Identity: 19%
PDB header:hydrolase regulator
Chain: A: PDB Molecule:phospholemman;
PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
Phyre2
| 13 |
|
PDB 3qnq chain D
Region: 414 - 450
Aligned: 37
Modelled: 37
Confidence: 10.1%
Identity: 16%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 14 |
|
PDB 2knc chain A
Region: 421 - 450
Aligned: 30
Modelled: 30
Confidence: 9.8%
Identity: 23%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 15 |
|
PDB 2klu chain A
Region: 422 - 447
Aligned: 26
Modelled: 26
Confidence: 6.1%
Identity: 19%
PDB header:immune system, membrane protein
Chain: A: PDB Molecule:t-cell surface glycoprotein cd4;
PDBTitle: nmr structure of the transmembrane and cytoplasmic domains2 of human cd4
Phyre2
| 16 |
|
PDB 2k2w chain A
Region: 433 - 449
Aligned: 17
Modelled: 17
Confidence: 5.7%
Identity: 18%
PDB header:cell cycle
Chain: A: PDB Molecule:recombination and dna repair protein;
PDBTitle: second brct domain of nbs1
Phyre2