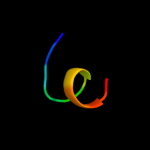| 1 |
|
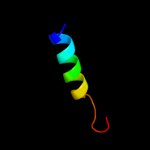
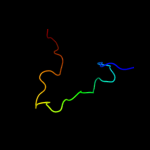
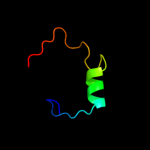
PDB 1kl9 chain A domain 1
Region: 81 - 97
Aligned: 16
Modelled: 17
Confidence: 37.4%
Identity: 13%
Fold: SAM domain-like
Superfamily: eIF2alpha middle domain-like
Family: eIF2alpha middle domain-like
Phyre2
| 2 |
|
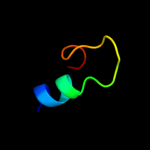
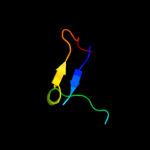
PDB 1q46 chain A domain 1
Region: 81 - 101
Aligned: 20
Modelled: 21
Confidence: 27.2%
Identity: 20%
Fold: SAM domain-like
Superfamily: eIF2alpha middle domain-like
Family: eIF2alpha middle domain-like
Phyre2
| 3 |
|
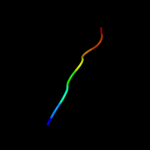
PDB 2vty chain A
Region: 84 - 103
Aligned: 20
Modelled: 20
Confidence: 18.8%
Identity: 25%
PDB header:apoptosis
Chain: A: PDB Molecule:protein f1;
PDBTitle: vaccinia virus anti-apoptotic f1l is a novel bcl-2-like2 domain swapped dimer
Phyre2
| 4 |
|
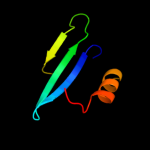
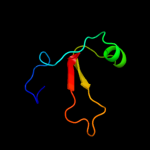
PDB 1u84 chain A
Region: 1 - 20
Aligned: 19
Modelled: 20
Confidence: 12.7%
Identity: 26%
Fold: YugE-like
Superfamily: YugE-like
Family: YugE-like
Phyre2
| 5 |
|
PDB 1fcd chain A domain 3
Region: 48 - 55
Aligned: 8
Modelled: 8
Confidence: 9.4%
Identity: 25%
Fold: CO dehydrogenase flavoprotein C-domain-like
Superfamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domain
Family: FAD/NAD-linked reductases, dimerisation (C-terminal) domain
Phyre2
| 6 |
|
PDB 3doa chain A
Region: 123 - 174
Aligned: 51
Modelled: 51
Confidence: 7.5%
Identity: 14%
PDB header:protein binding
Chain: A: PDB Molecule:fibrinogen binding protein;
PDBTitle: the crystal structure of the fibrinogen binding protein from2 staphylococcus aureus
Phyre2
| 7 |
|
PDB 2q8a chain A
Region: 130 - 163
Aligned: 34
Modelled: 34
Confidence: 7.4%
Identity: 12%
PDB header:immune system
Chain: A: PDB Molecule:apical membrane antigen 1;
PDBTitle: structure of the malaria antigen ama1 in complex with a growth-2 inhibitory antibody
Phyre2
| 8 |
|
PDB 1sqw chain A domain 1
Region: 43 - 68
Aligned: 26
Modelled: 26
Confidence: 7.0%
Identity: 23%
Fold: PUA domain-like
Superfamily: PUA domain-like
Family: PUA domain
Phyre2
| 9 |
|
PDB 1cz5 chain A domain 1
Region: 135 - 151
Aligned: 17
Modelled: 17
Confidence: 6.1%
Identity: 18%
Fold: Double psi beta-barrel
Superfamily: ADC-like
Family: Cdc48 N-terminal domain-like
Phyre2
| 10 |
|
PDB 1mv8 chain A domain 1
Region: 49 - 58
Aligned: 10
Modelled: 10
Confidence: 5.9%
Identity: 30%
Fold: 6-phosphogluconate dehydrogenase C-terminal domain-like
Superfamily: 6-phosphogluconate dehydrogenase C-terminal domain-like
Family: UDP-glucose/GDP-mannose dehydrogenase dimerisation domain
Phyre2
| 11 |
|
PDB 1dzf chain A domain 2
Region: 82 - 130
Aligned: 49
Modelled: 49
Confidence: 5.7%
Identity: 24%
Fold: RPB5-like RNA polymerase subunit
Superfamily: RPB5-like RNA polymerase subunit
Family: RPB5
Phyre2
| 12 |
|
PDB 1tp6 chain A
Region: 12 - 45
Aligned: 31
Modelled: 34
Confidence: 5.6%
Identity: 16%
Fold: Cystatin-like
Superfamily: NTF2-like
Family: PA1314-like
Phyre2
| 13 |
|
PDB 1z67 chain A domain 1
Region: 86 - 104
Aligned: 18
Modelled: 19
Confidence: 5.6%
Identity: 44%
Fold: YidB-like
Superfamily: YidB-like
Family: YidB-like
Phyre2