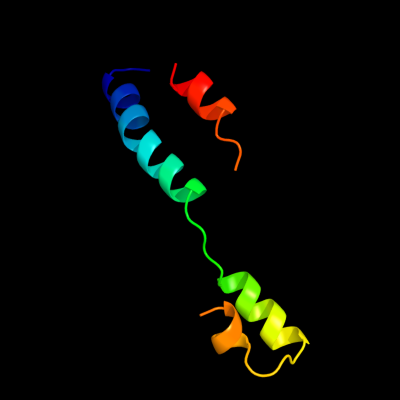
1 c3chtA_
48.4
18
PDB header: oxidoreductaseChain: A: PDB Molecule: p-aminobenzoate n-oxygenase;PDBTitle: crystal structure of di-iron aurf with partially bound ligand
2 c3gzxB_
47.9
13
PDB header: oxidoreductaseChain: B: PDB Molecule: biphenyl dioxygenase subunit beta;PDBTitle: crystal structure of the biphenyl dioxygenase in complex with biphenyl2 from comamonas testosteroni sp. strain b-356
3 d1ktdb2
41.7
31
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain4 d1i3rb2
40.0
38
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain5 d1klub2
38.4
62
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain6 d2pxyd2
38.4
23
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain7 d2iadb2
37.2
38
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain8 d1d5mb2
36.7
50
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain9 d1iakb2
36.4
31
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain10 d1bx2b2
36.1
62
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain11 d1fv1b2
34.2
62
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain12 d1s9vb2
33.8
38
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain13 d1uvqb2
32.8
31
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain14 d3c5zd2
32.7
31
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain15 d1mujb2
32.7
31
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain16 d2nnab2
32.4
31
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain17 d1es0b2
32.1
31
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain18 d1fngb2
32.0
38
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain19 d1dd4c_
30.1
18
Fold: Ribosomal protein L7/12, oligomerisation (N-terminal) domainSuperfamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domainFamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domain20 d1a6ab2
28.8
54
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain21 d1w7ca2
not modelled
27.4
17
Fold: Cystatin-likeSuperfamily: Amine oxidase N-terminal regionFamily: Amine oxidase N-terminal region22 d1ulib_
not modelled
23.6
14
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Ring hydroxylating beta subunit23 d1z3xa1
not modelled
22.5
19
Fold: alpha-alpha superhelixSuperfamily: ARM repeatFamily: GUN4-associated domain24 c2gbxF_
not modelled
21.5
14
PDB header: oxidoreductaseChain: F: PDB Molecule: biphenyl 2,3-dioxygenase beta subunit;PDBTitle: crystal structure of biphenyl 2,3-dioxygenase from sphingomonas2 yanoikuyae b1 bound to biphenyl
25 d1kkxa_
not modelled
20.4
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: ARID-likeFamily: ARID domain26 c1dd3C_
not modelled
19.7
19
PDB header: ribosomeChain: C: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: crystal structure of ribosomal protein l12 from thermotoga maritima
27 c1dd3D_
not modelled
19.7
19
PDB header: ribosomeChain: D: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: crystal structure of ribosomal protein l12 from thermotoga maritima
28 d1dd4d_
not modelled
19.0
19
Fold: Ribosomal protein L7/12, oligomerisation (N-terminal) domainSuperfamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domainFamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domain29 d1g4ma1
not modelled
18.8
24
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Arrestin/Vps26-like30 c2ib1A_
not modelled
18.7
15
PDB header: apoptosisChain: A: PDB Molecule: death domain containing membrane protein nradd;PDBTitle: solution structure of p45 death domain
31 c2kkvA_
not modelled
17.9
17
PDB header: dna binding proteinChain: A: PDB Molecule: integrase;PDBTitle: solution nmr structure of an integrase domain from protein2 spa4288 from salmonella enterica, northeast structural3 genomics consortium target slr105h
32 c2hl7A_
not modelled
17.7
21
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c-type biogenesis protein ccmh;PDBTitle: crystal structure of the periplasmic domain of ccmh from pseudomonas2 aeruginosa
33 c2k9sA_
not modelled
17.2
17
PDB header: transcriptionChain: A: PDB Molecule: arabinose operon regulatory protein;PDBTitle: solution structure of dna binding domain of e. coli arac
34 c2kj8A_
not modelled
16.9
16
PDB header: dna binding proteinChain: A: PDB Molecule: putative prophage cps-53 integrase;PDBTitle: nmr structure of fragment 87-196 from the putative phage2 integrase ints of e. coli: northeast structural genomics3 consortium target er652a, psi-2
35 c1nohB_
not modelled
16.5
27
PDB header: viral proteinChain: B: PDB Molecule: head morphogenesis protein;PDBTitle: the structure of bacteriophage phi29 scaffolding protein2 gp7 after prohead assembly
36 d2coba1
not modelled
16.5
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Psq domain37 c1bl0A_
not modelled
16.4
14
PDB header: transcription/dnaChain: A: PDB Molecule: protein (multiple antibiotic resistance protein);PDBTitle: multiple antibiotic resistance protein (mara)/dna complex
38 c2lboA_
not modelled
16.1
25
PDB header: cell adhesionChain: A: PDB Molecule: microneme protein 3;PDBTitle: eimeria tenella microneme protein 3 mar_b domain
39 c2kkpA_
not modelled
15.5
16
PDB header: dna binding proteinChain: A: PDB Molecule: phage integrase;PDBTitle: solution nmr structure of the phage integrase sam-like2 domain from moth 1796 from moorella thermoacetica.3 northeast structural genomics consortium target mtr39k4 (residues 64-171).
40 c2wx3A_
not modelled
14.8
46
PDB header: structural proteinChain: A: PDB Molecule: mrna-decapping enzyme 1a;PDBTitle: asymmetric trimer of the human dcp1a c-terminal domain
41 c3lysC_
not modelled
13.7
8
PDB header: recombinationChain: C: PDB Molecule: prophage pi2 protein 01, integrase;PDBTitle: crystal structure of the n-terminal domain of the prophage2 pi2 protein 01 (integrase) from lactococcus lactis,3 northeast structural genomics consortium target kr124f
42 d2g2ub1
not modelled
13.7
25
Fold: BLIP-likeSuperfamily: beta-lactamase-inhibitor protein, BLIPFamily: beta-lactamase-inhibitor protein, BLIP43 c2kw0A_
not modelled
13.6
21
PDB header: oxidoreductaseChain: A: PDB Molecule: ccmh protein;PDBTitle: solution structure of n-terminal domain of ccmh from escherichia.coli
44 c1giyJ_
not modelled
13.2
20
PDB header: ribosomeChain: J: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: crystal structure of the ribosome at 5.5 a resolution. this2 file, 1giy, contains the 50s ribosome subunit. the 30s3 ribosome subunit, three trna, and mrna molecules are in the4 file 1gix
45 c3ahrA_
not modelled
13.2
26
PDB header: oxidoreductaseChain: A: PDB Molecule: ero1-like protein alpha;PDBTitle: inactive human ero1
46 c2khqA_
not modelled
12.8
11
PDB header: dna binding proteinChain: A: PDB Molecule: integrase;PDBTitle: solution nmr structure of a phage integrase ssp19472 fragment 59-159 from staphylococcus saprophyticus,3 northeast structural genomics consortium target syr103b
47 c3oa8B_
not modelled
12.7
19
PDB header: heme-binding protein/heme-binding proteiChain: B: PDB Molecule: soxx;PDBTitle: diheme soxax
48 c3ol4B_
not modelled
12.6
27
PDB header: unknown functionChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of a putative uncharacterized protein from2 mycobacterium smegmatis, an ortholog of rv0543c
49 c3btpA_
not modelled
12.5
25
PDB header: dna binding protein, chaperoneChain: A: PDB Molecule: single-strand dna-binding protein;PDBTitle: crystal structure of agrobacterium tumefaciens vire2 in complex with2 its chaperone vire1: a novel fold and implications for dna binding
50 d1usta_
not modelled
12.0
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Linker histone H1/H551 c2kj5A_
not modelled
11.9
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: phage integrase;PDBTitle: solution nmr structure of a domain from a putative phage2 integrase protein nmul_a0064 from nitrosospira multiformis,3 northeast structural genomics consortium target nmr46c
52 c2kj9A_
not modelled
11.9
11
PDB header: dna binding proteinChain: A: PDB Molecule: integrase;PDBTitle: nmr structure of intb phage-integrase-like protein fragment2 90-199 from erwinia carotova subsp. atroseptica: northeast3 structural genomics consortium target ewr217e
53 c2nyxB_
not modelled
11.7
36
PDB header: transcriptionChain: B: PDB Molecule: probable transcriptional regulatory protein, rv1404;PDBTitle: crystal structure of rv1404 from mycobacterium tuberculosis
54 c2noxP_
not modelled
11.7
19
PDB header: oxidoreductaseChain: P: PDB Molecule: tryptophan 2,3-dioxygenase;PDBTitle: crystal structure of tryptophan 2,3-dioxygenase from ralstonia2 metallidurans
55 d3ebya1
not modelled
11.6
14
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Ring hydroxylating beta subunit56 c2dxbR_
not modelled
11.5
26
PDB header: hydrolaseChain: R: PDB Molecule: thiocyanate hydrolase subunit gamma;PDBTitle: recombinant thiocyanate hydrolase comprising partially-modified cobalt2 centers
57 c3pstA_
not modelled
11.4
27
PDB header: nuclear proteinChain: A: PDB Molecule: protein doa1;PDBTitle: crystal structure of pul and pfu(mutate) domain
58 c2ee7A_
not modelled
11.0
11
PDB header: structural proteinChain: A: PDB Molecule: sperm flagellar protein 1;PDBTitle: solution structure of the ch domain from human sperm2 flagellar protein 1
59 c2d96A_
not modelled
10.8
19
PDB header: transcriptionChain: A: PDB Molecule: nuclear factor nf-kappa-b p100 subunit;PDBTitle: solution structure of the death domain of nuclear factor nf-2 kappa-b p100
60 d1uzxa_
not modelled
10.8
25
Fold: UBC-likeSuperfamily: UBC-likeFamily: UEV domain61 c3m5bA_
not modelled
10.7
10
PDB header: transcriptionChain: A: PDB Molecule: zinc finger and btb domain-containing protein 32;PDBTitle: crystal structure of the btb domain from fazf/zbtb32
62 c2z3xC_
not modelled
10.4
29
PDB header: dna binding protein/dnaChain: C: PDB Molecule: small, acid-soluble spore protein c;PDBTitle: structure of a protein-dna complex essential for dna2 protection in spore of bacillus species
63 d1dd3a1
not modelled
10.3
20
Fold: Ribosomal protein L7/12, oligomerisation (N-terminal) domainSuperfamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domainFamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domain64 c2ld7A_
not modelled
10.0
24
PDB header: transcriptionChain: A: PDB Molecule: histone deacetylase complex subunit sap30;PDBTitle: solution structure of the msin3a pah3-sap30 sid complex
65 d1iufa1
not modelled
9.9
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Centromere-binding66 d1ig6a_
not modelled
9.9
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: ARID-likeFamily: ARID domain67 c2jhdA_
not modelled
9.7
11
PDB header: cell adhesionChain: A: PDB Molecule: micronemal protein 1;PDBTitle: crystal structure of toxoplasma gondii micronemal protein 12 bound to 3'-sialyl-n-acetyllactosamine
68 d1s7oa_
not modelled
9.3
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: YlxM/p13-like69 c2jxjA_
not modelled
9.2
27
PDB header: oxidoreductaseChain: A: PDB Molecule: histone demethylase jarid1a;PDBTitle: nmr structure of the arid domain from the histone h3k42 demethylase rbp2
70 c2khvA_
not modelled
8.9
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: phage integrase;PDBTitle: solution nmr structure of protein nmul_a0922 from2 nitrosospira multiformis. northeast structural genomics3 consortium target nmr38b.
71 d1h9xa1
not modelled
8.7
28
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: N-terminal (heme c) domain of cytochrome cd1-nitrite reductase72 c3lsgD_
not modelled
8.6
14
PDB header: transcription regulatorChain: D: PDB Molecule: two-component response regulator yesn;PDBTitle: the crystal structure of the c-terminal domain of the two-2 component response regulator yesn from fusobacterium3 nucleatum subsp. nucleatum atcc 25586
73 c3oioA_
not modelled
8.6
21
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator (arac-type dna-binding domain-PDBTitle: crystal structure of transcriptional regulator (arac-type dna-binding2 domain-containing proteins) from chromobacterium violaceum
74 c2krfB_
not modelled
8.5
10
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulatory protein coma;PDBTitle: nmr solution structure of the dna binding domain of competence protein2 a
75 d1zu1a1
not modelled
8.5
14
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: HkH motif-containing C2H2 finger76 d2b1xb1
not modelled
8.5
7
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Ring hydroxylating beta subunit77 d1hlma_
not modelled
8.4
14
Fold: Globin-likeSuperfamily: Globin-likeFamily: Globins78 c2ou2A_
not modelled
8.4
15
PDB header: transferaseChain: A: PDB Molecule: histone acetyltransferase htatip;PDBTitle: acetyltransferase domain of human hiv-1 tat interacting2 protein, 60kda, isoform 3
79 c2jnsA_
not modelled
8.4
13
PDB header: unknown functionChain: A: PDB Molecule: bromodomain-containing protein 4;PDBTitle: solution structure of the bromodomain-containing protein 42 et domain
80 d1dzfa1
not modelled
8.4
9
Fold: Restriction endonuclease-likeSuperfamily: Eukaryotic RPB5 N-terminal domainFamily: Eukaryotic RPB5 N-terminal domain81 d1wqlb1
not modelled
8.3
18
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Ring hydroxylating beta subunit82 d2ozua1
not modelled
8.2
21
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-acetyl transferase, NAT83 c2ys9A_
not modelled
8.2
28
PDB header: transcriptionChain: A: PDB Molecule: homeobox and leucine zipper protein homez;PDBTitle: structure of the third homeodomain from the human homeobox2 and leucine zipper protein, homez
84 d1yqaa1
not modelled
8.1
9
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Linker histone H1/H585 d1kv9a1
not modelled
8.1
20
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinoprotein alcohol dehydrogenase, C-terminal domain86 d1ijwc_
not modelled
8.1
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain87 c3h9xB_
not modelled
8.0
29
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein pspto_3016;PDBTitle: crystal structure of the pspto_3016 protein from2 pseudomonas syringae, northeast structural genomics3 consortium target psr293
88 c2obuA_
not modelled
7.8
28
PDB header: hormone/growth factorChain: A: PDB Molecule: gastric inhibitory polypeptide;PDBTitle: solution structure of gip in tfe/water
89 c3ixzA_
not modelled
7.8
18
PDB header: hydrolaseChain: A: PDB Molecule: potassium-transporting atpase alpha;PDBTitle: pig gastric h+/k+-atpase complexed with aluminium fluoride
90 c3l3fX_
not modelled
7.8
39
PDB header: protein bindingChain: X: PDB Molecule: protein doa1;PDBTitle: crystal structure of a pfu-pul domain pair of saccharomyces cerevisiae2 doa1/ufd3
91 d3ejva1
not modelled
7.7
9
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: BaiE/LinA-like92 c1iijA_
not modelled
7.7
33
PDB header: signaling proteinChain: A: PDB Molecule: erbb-2 receptor protein-tyrosine kinase;PDBTitle: solution structure of the neu/erbb-2 membrane spanning2 segment
93 d1eb7a2
not modelled
7.5
15
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Di-heme cytochrome c peroxidase94 d1wmga_
not modelled
7.4
11
Fold: DEATH domainSuperfamily: DEATH domainFamily: DEATH domain, DD95 c2kw7A_
not modelled
7.3
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved domain protein;PDBTitle: solution nmr structure of the n-terminal domain of protein pg_03612 from p.gingivalis, northeast structural genomics consortium target3 pgr37a
96 c2c1dB_
not modelled
7.3
22
PDB header: oxidoreductaseChain: B: PDB Molecule: soxx;PDBTitle: crystal structure of soxxa from p. pantotrophus
97 d3e99a1
not modelled
7.2
7
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Ring hydroxylating beta subunit98 c2eqxA_
not modelled
7.2
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: kelch repeat and btb domain-containing protein 4;PDBTitle: solution structure of the back domain of kelch repeat and2 btb domain-containing protein 4
99 d1hcra_
not modelled
7.0
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain