| 1 |
|
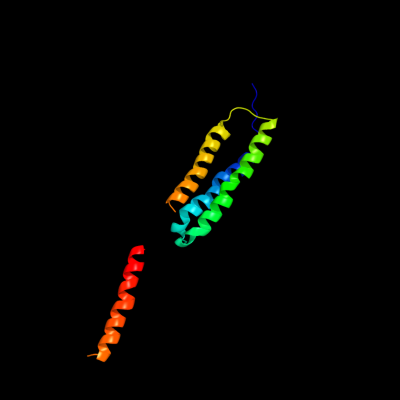
PDB 2bg9 chain B
Region: 4 - 137
Aligned: 118
Modelled: 119
Confidence: 63.4%
Identity: 14%
PDB header:ion channel/receptor
Chain: B: PDB Molecule:acetylcholine receptor protein, beta chain;
PDBTitle: refined structure of the nicotinic acetylcholine receptor2 at 4a resolution.
Phyre2
| 2 |
|
PDB 2knc chain B
Region: 105 - 145
Aligned: 39
Modelled: 41
Confidence: 60.6%
Identity: 10%
PDB header:cell adhesion
Chain: B: PDB Molecule:integrin beta-3;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 3 |
|
PDB 1vpu chain A
Region: 121 - 145
Aligned: 25
Modelled: 25
Confidence: 51.4%
Identity: 24%
Fold: HIV-1 VPU cytoplasmic domain
Superfamily: HIV-1 VPU cytoplasmic domain
Family: HIV-1 VPU cytoplasmic domain
Phyre2
| 4 |
|
PDB 3ria chain C
Region: 4 - 95
Aligned: 92
Modelled: 92
Confidence: 44.1%
Identity: 7%
PDB header:transport protein/immune system
Chain: C: PDB Molecule:avermectin-sensitive glutamate-gated chloride channel glucl
PDBTitle: c. elegans glutamate-gated chloride channel (glucl) in complex with2 fab, ivermectin and iodide.
Phyre2
| 5 |
|
PDB 1kqf chain C
Region: 2 - 119
Aligned: 118
Modelled: 118
Confidence: 43.1%
Identity: 14%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Formate dehydrogenase N, cytochrome (gamma) subunit
Phyre2
| 6 |
|
PDB 2bg9 chain A
Region: 4 - 92
Aligned: 88
Modelled: 89
Confidence: 32.4%
Identity: 14%
PDB header:ion channel/receptor
Chain: A: PDB Molecule:acetylcholine receptor protein, alpha chain;
PDBTitle: refined structure of the nicotinic acetylcholine receptor2 at 4a resolution.
Phyre2
| 7 |
|
PDB 2bg9 chain C
Region: 4 - 142
Aligned: 123
Modelled: 124
Confidence: 30.7%
Identity: 11%
PDB header:ion channel/receptor
Chain: C: PDB Molecule:acetylcholine receptor protein, delta chain;
PDBTitle: refined structure of the nicotinic acetylcholine receptor2 at 4a resolution.
Phyre2
| 8 |
|
PDB 3eam chain B
Region: 4 - 95
Aligned: 90
Modelled: 92
Confidence: 26.6%
Identity: 9%
PDB header:membrane protein, transport protein
Chain: B: PDB Molecule:glr4197 protein;
PDBTitle: an open-pore structure of a bacterial pentameric ligand-2 gated ion channel
Phyre2
| 9 |
|
PDB 1m8o chain B
Region: 121 - 145
Aligned: 25
Modelled: 25
Confidence: 23.4%
Identity: 12%
PDB header:membrane protein
Chain: B: PDB Molecule:platele integrin beta3 subunit: cytoplasmic
PDBTitle: platelet integrin alfaiib-beta3 cytoplasmic domain
Phyre2
| 10 |
|
PDB 1s4x chain A
Region: 121 - 145
Aligned: 25
Modelled: 25
Confidence: 22.3%
Identity: 12%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin beta-3;
PDBTitle: nmr structure of the integrin b3 cytoplasmic domain in dpc2 micelles
Phyre2
| 11 |
|
PDB 2ksr chain A
Region: 6 - 92
Aligned: 86
Modelled: 87
Confidence: 18.7%
Identity: 16%
PDB header:membrane protein
Chain: A: PDB Molecule:neuronal acetylcholine receptor subunit beta-2;
PDBTitle: nmr structures of tm domain of the n-acetylcholine receptor b2 subunit
Phyre2
| 12 |
|
PDB 3g9w chain C
Region: 124 - 145
Aligned: 22
Modelled: 22
Confidence: 14.6%
Identity: 14%
PDB header:cell adhesion
Chain: C: PDB Molecule:integrin beta-1d;
PDBTitle: crystal structure of talin2 f2-f3 in complex with the integrin beta1d2 cytoplasmic tail
Phyre2
| 13 |
|
PDB 1q90 chain L
Region: 13 - 32
Aligned: 20
Modelled: 20
Confidence: 11.1%
Identity: 30%
PDB header:photosynthesis
Chain: L: PDB Molecule:cytochrome b6f complex subunit petl;
PDBTitle: structure of the cytochrome b6f (plastohydroquinone : plastocyanin2 oxidoreductase) from chlamydomonas reinhardtii
Phyre2
| 14 |
|
PDB 1q90 chain L
Region: 13 - 32
Aligned: 20
Modelled: 20
Confidence: 11.1%
Identity: 30%
Fold: Single transmembrane helix
Superfamily: PetL subunit of the cytochrome b6f complex
Family: PetL subunit of the cytochrome b6f complex
Phyre2
| 15 |
|
PDB 1j46 chain A
Region: 1 - 16
Aligned: 16
Modelled: 16
Confidence: 9.6%
Identity: 25%
Fold: HMG-box
Superfamily: HMG-box
Family: HMG-box
Phyre2
| 16 |
|
PDB 1kuz chain B
Region: 124 - 144
Aligned: 21
Modelled: 21
Confidence: 9.2%
Identity: 14%
PDB header:cell adhesion
Chain: B: PDB Molecule:integrin beta-3;
PDBTitle: solution structure of the membrane proximal regions of2 alpha-iib and beta-3 integrins
Phyre2
| 17 |
|
PDB 1uaz chain A
Region: 4 - 137
Aligned: 134
Modelled: 134
Confidence: 8.1%
Identity: 13%
Fold: Family A G protein-coupled receptor-like
Superfamily: Family A G protein-coupled receptor-like
Family: Bacteriorhodopsin-like
Phyre2
| 18 |
|
PDB 2bg9 chain E
Region: 4 - 137
Aligned: 126
Modelled: 126
Confidence: 7.9%
Identity: 10%
PDB header:ion channel/receptor
Chain: E: PDB Molecule:acetylcholine receptor protein, gamma chain;
PDBTitle: refined structure of the nicotinic acetylcholine receptor2 at 4a resolution.
Phyre2
| 19 |
|
PDB 3omy chain B
Region: 61 - 97
Aligned: 31
Modelled: 37
Confidence: 6.6%
Identity: 19%
PDB header:dna binding protein
Chain: B: PDB Molecule:protein tram;
PDBTitle: crystal structure of the ped208 tram n-terminal domain
Phyre2
| 20 |
|
PDB 1i7n chain A domain 1
Region: 122 - 145
Aligned: 24
Modelled: 24
Confidence: 6.1%
Identity: 33%
Fold: PreATP-grasp domain
Superfamily: PreATP-grasp domain
Family: Synapsin domain
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|
| 24 |
|