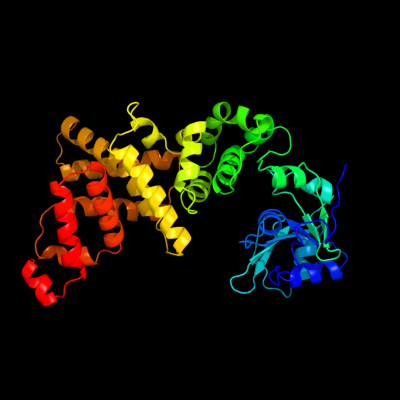
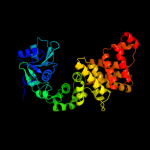
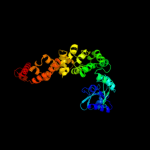
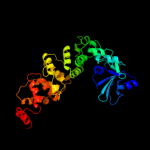
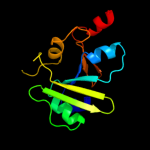
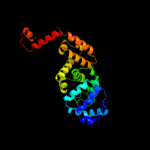
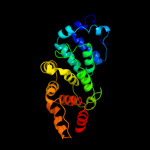
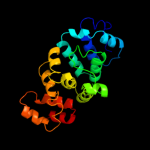
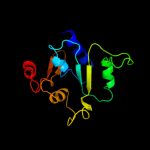
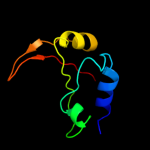
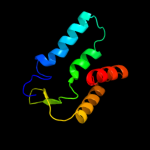
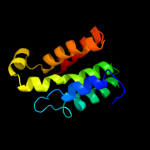
1 c3aqnA_
100.0
100
PDB header: transferaseChain: A: PDB Molecule: poly(a) polymerase;PDBTitle: complex structure of bacterial protein (apo form ii)
2 c1miyB_
100.0
28
PDB header: translation, transferaseChain: B: PDB Molecule: trna cca-adding enzyme;PDBTitle: crystal structure of bacillus stearothermophilus cca-adding enzyme in2 complex with ctp
3 c1vfgB_
100.0
24
PDB header: transferase/rnaChain: B: PDB Molecule: poly a polymerase;PDBTitle: crystal structure of trna nucleotidyltransferase complexed2 with a primer trna and an incoming atp analog
4 c3h37B_
100.0
21
PDB header: transferaseChain: B: PDB Molecule: trna nucleotidyl transferase-related protein;PDBTitle: the structure of cca-adding enzyme apo form i
5 c1ou5A_
100.0
26
PDB header: translation, transferaseChain: A: PDB Molecule: trna cca-adding enzyme;PDBTitle: crystal structure of human cca-adding enzyme
6 d1miwa2
100.0
38
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Poly A polymerase head domain-like7 d1ou5a2
100.0
35
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Poly A polymerase head domain-like8 d1vfga2
100.0
40
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Poly A polymerase head domain-like9 d1miwa1
100.0
21
Fold: Poly A polymerase C-terminal region-likeSuperfamily: Poly A polymerase C-terminal region-likeFamily: Poly A polymerase C-terminal region-like10 d1vfga1
100.0
18
Fold: Poly A polymerase C-terminal region-likeSuperfamily: Poly A polymerase C-terminal region-likeFamily: Poly A polymerase C-terminal region-like11 d1ou5a1
100.0
19
Fold: Poly A polymerase C-terminal region-likeSuperfamily: Poly A polymerase C-terminal region-likeFamily: Poly A polymerase C-terminal region-like12 c2la3A_
96.7
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: the nmr structure of the protein np_344798.1 reveals a cca-adding2 enzyme head domain
13 d2fcla1
91.6
22
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: TM1012-like14 d2qgsa1
86.8
10
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain15 d2pq7a1
80.8
7
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain16 d1r89a2
69.0
20
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Archaeal tRNA CCA-adding enzyme catalytic domain17 d3djba1
63.8
14
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain18 d1y81a1
58.2
23
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain19 d2d59a1
46.9
23
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain20 d1jmsa4
43.2
19
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: DNA polymerase beta-like21 d1iowa1
not modelled
39.9
20
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: D-Alanine ligase N-terminal domain22 c2q14A_
not modelled
38.4
7
PDB header: hydrolaseChain: A: PDB Molecule: phosphohydrolase;PDBTitle: crystal structure of phosphohydrolase (bt4208) from bacteroides2 thetaiotaomicron vpi-5482 at 2.20 a resolution
23 c3mdoB_
not modelled
34.9
32
PDB header: ligaseChain: B: PDB Molecule: putative phosphoribosylformylglycinamidine cyclo-ligase;PDBTitle: crystal structure of a putative phosphoribosylformylglycinamidine2 cyclo-ligase (bdi_2101) from parabacteroides distasonis atcc 8503 at3 1.91 a resolution
24 d1b7go1
not modelled
34.9
27
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain25 d2fmpa3
not modelled
33.0
26
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: DNA polymerase beta-like26 c3skdA_
not modelled
31.0
11
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein tthb187;PDBTitle: crystal structure of the thermus thermophilus cas3 hd domain in the2 presence of ni2+
27 c3fpnA_
not modelled
30.3
18
PDB header: dna binding proteinChain: A: PDB Molecule: geobacillus stearothermophilus uvra interactionPDBTitle: crystal structure of uvra-uvrb interaction domains
28 d1n57a_
not modelled
29.2
17
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: DJ-1/PfpI29 c2duwA_
not modelled
28.9
21
PDB header: ligand binding proteinChain: A: PDB Molecule: putative coa-binding protein;PDBTitle: solution structure of putative coa-binding protein of2 klebsiella pneumoniae
30 c1x60A_
not modelled
28.5
30
PDB header: hydrolaseChain: A: PDB Molecule: sporulation-specific n-acetylmuramoyl-l-alaninePDBTitle: solution structure of the peptidoglycan binding domain of2 b. subtilis cell wall lytic enzyme cwlc
31 c2pcnA_
not modelled
28.5
18
PDB header: transferaseChain: A: PDB Molecule: s-adenosylmethionine:2-demethylmenaquinonePDBTitle: crystal structure of s-adenosylmethionine: 2-dimethylmenaquinone2 methyltransferase (gk_1813) from geobacillus kaustophilus hta426
32 c2ze5A_
not modelled
28.0
13
PDB header: transferaseChain: A: PDB Molecule: isopentenyl transferase;PDBTitle: crystal structure of adenosine phosphate-isopentenyltransferase
33 d2bcqa3
not modelled
27.6
23
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: DNA polymerase beta-like34 c3k4iC_
not modelled
27.0
18
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of uncharacterized protein pspto_3204 from2 pseudomonas syringae pv. tomato str. dc3000
35 d2bdua1
not modelled
26.9
21
Fold: HAD-likeSuperfamily: HAD-likeFamily: Pyrimidine 5'-nucleotidase (UMPH-1)36 c3c8oB_
not modelled
24.9
32
PDB header: hydrolase regulatorChain: B: PDB Molecule: regulator of ribonuclease activity a;PDBTitle: the crystal structure of rraa from pao1
37 d1vi4a_
not modelled
24.9
27
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: RraA-likeFamily: RraA-like38 d1no5a_
not modelled
24.0
25
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Catalytic subunit of bi-partite nucleotidyltransferase39 d1clia2
not modelled
23.9
32
Fold: PurM C-terminal domain-likeSuperfamily: PurM C-terminal domain-likeFamily: PurM C-terminal domain-like40 d1j3la_
not modelled
23.8
32
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: RraA-likeFamily: RraA-like41 d2vana2
not modelled
23.5
26
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: DNA polymerase beta-like42 d1q5xa_
not modelled
23.3
18
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: RraA-likeFamily: RraA-like43 d1iuka_
not modelled
23.3
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain44 c3m5fA_
not modelled
23.2
10
PDB header: hydrolaseChain: A: PDB Molecule: metal dependent phosphohydrolase;PDBTitle: structure of mj0384, a cas3 protein from methanocaldococcus jannaschii
45 c1nxjA_
not modelled
23.1
27
PDB header: unknown functionChain: A: PDB Molecule: probable s-adenosylmethionine:2-PDBTitle: structure of rv3853 from mycobacterium tuberculosis
46 d1nxja_
not modelled
23.1
27
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: RraA-likeFamily: RraA-like47 c2c5qE_
not modelled
22.7
23
PDB header: structural genomics,unknown functionChain: E: PDB Molecule: rraa-like protein yer010c;PDBTitle: crystal structure of yeast yer010cp
48 c3a8tA_
not modelled
22.6
15
PDB header: transferaseChain: A: PDB Molecule: adenylate isopentenyltransferase;PDBTitle: plant adenylate isopentenyltransferase in complex with atp
49 d1wota_
not modelled
22.5
12
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Catalytic subunit of bi-partite nucleotidyltransferase50 d2vkqa1
not modelled
22.2
19
Fold: HAD-likeSuperfamily: HAD-likeFamily: Pyrimidine 5'-nucleotidase (UMPH-1)51 d1yt8a4
not modelled
21.9
20
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)52 c3d54I_
not modelled
21.3
22
PDB header: ligaseChain: I: PDB Molecule: phosphoribosylformylglycinamidine synthase ii;PDBTitle: stucture of purlqs from thermotoga maritima
53 c3g5jA_
not modelled
21.1
22
PDB header: nucleotide binding proteinChain: A: PDB Molecule: putative atp/gtp binding protein;PDBTitle: crystal structure of n-terminal domain of putative atp/gtp binding2 protein from clostridium difficile 630
54 c2w9mB_
not modelled
21.0
19
PDB header: dna replicationChain: B: PDB Molecule: polymerase x;PDBTitle: structure of family x dna polymerase from deinococcus2 radiodurans
55 d2fuea1
not modelled
20.0
14
Fold: HAD-likeSuperfamily: HAD-likeFamily: Predicted hydrolases Cof56 c3exaD_
not modelled
19.9
18
PDB header: transferaseChain: D: PDB Molecule: trna delta(2)-isopentenylpyrophosphatePDBTitle: crystal structure of the full-length trna2 isopentenylpyrophosphate transferase (bh2366) from3 bacillus halodurans, northeast structural genomics4 consortium target bhr41.
57 d1wpga2
not modelled
19.4
13
Fold: HAD-likeSuperfamily: HAD-likeFamily: Meta-cation ATPase, catalytic domain P58 d1vk3a1
not modelled
18.7
53
Fold: Bacillus chorismate mutase-likeSuperfamily: PurM N-terminal domain-likeFamily: PurM N-terminal domain-like59 c3d3qB_
not modelled
18.1
15
PDB header: transferaseChain: B: PDB Molecule: trna delta(2)-isopentenylpyrophosphatePDBTitle: crystal structure of trna delta(2)-isopentenylpyrophosphate2 transferase (se0981) from staphylococcus epidermidis.3 northeast structural genomics consortium target ser100
60 c3ib6B_
not modelled
17.6
12
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an uncharacterized protein from listeria2 monocytogenes serotype 4b
61 d2heka1
not modelled
17.4
17
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain62 c2zodB_
not modelled
16.7
18
PDB header: transferaseChain: B: PDB Molecule: selenide, water dikinase;PDBTitle: crystal structure of selenophosphate synthetase from2 aquifex aeolicus
63 c2cx9C_
not modelled
16.6
16
PDB header: oxidoreductaseChain: C: PDB Molecule: acyl-coa dehydrogenase;PDBTitle: crystal structure of acyl-coa dehydrogenase
64 c3kizA_
not modelled
16.2
24
PDB header: ligaseChain: A: PDB Molecule: phosphoribosylformylglycinamidine cyclo-ligase;PDBTitle: crystal structure of putative phosphoribosylformylglycinamidine cyclo-2 ligase (yp_676759.1) from cytophaga hutchinsonii atcc 33406 at 1.50 a3 resolution
65 c3crqA_
not modelled
16.0
22
PDB header: transferaseChain: A: PDB Molecule: trna delta(2)-isopentenylpyrophosphatePDBTitle: structure of trna dimethylallyltransferase: rna2 modification through a channel
66 c2yxbA_
not modelled
16.0
25
PDB header: isomeraseChain: A: PDB Molecule: coenzyme b12-dependent mutase;PDBTitle: crystal structure of the methylmalonyl-coa mutase alpha-subunit from2 aeropyrum pernix
67 c1wv9B_
not modelled
15.8
32
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: rhodanese homolog tt1651;PDBTitle: crystal structure of rhodanese homolog tt1651 from an2 extremely thermophilic bacterium thermus thermophilus hb8
68 c1sz1A_
not modelled
15.6
20
PDB header: transferase/rnaChain: A: PDB Molecule: trna nucleotidyltransferase;PDBTitle: mechanism of cca-adding enzymes specificity revealed by crystal2 structures of ternary complexes
69 c3nojA_
not modelled
15.6
36
PDB header: lyaseChain: A: PDB Molecule: 4-carboxy-4-hydroxy-2-oxoadipate aldolase/oxaloacetatePDBTitle: the structure of hmg/cha aldolase from the protocatechuate degradation2 pathway of pseudomonas putida
70 c2pfrB_
not modelled
15.5
22
PDB header: transferaseChain: B: PDB Molecule: arylamine n-acetyltransferase 2;PDBTitle: human n-acetyltransferase 2
71 d2b8ea1
not modelled
15.5
21
Fold: HAD-likeSuperfamily: HAD-likeFamily: Meta-cation ATPase, catalytic domain P72 c3d9wA_
not modelled
15.0
25
PDB header: transferaseChain: A: PDB Molecule: putative acetyltransferase;PDBTitle: crystal structure analysis of nocardia farcinica arylamine2 n-acetyltransferase
73 d1p49a_
not modelled
14.6
13
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Arylsulfatase74 d1w4ta1
not modelled
13.7
40
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Arylamine N-acetyltransferase75 c2crlA_
not modelled
12.7
32
PDB header: chaperoneChain: A: PDB Molecule: copper chaperone for superoxide dismutase;PDBTitle: the apo form of hma domain of copper chaperone for2 superoxide dismutase
76 c3ff4A_
not modelled
12.3
32
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of uncharacterized protein chu_1412
77 c3lnbA_
not modelled
12.1
33
PDB header: transferaseChain: A: PDB Molecule: n-acetyltransferase family protein;PDBTitle: crystal structure analysis of arylamine n-acetyltransferase c from2 bacillus anthracis
78 d1r9la_
not modelled
12.0
19
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like79 d1zpda1
not modelled
11.9
16
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain80 c3ix9B_
not modelled
11.9
19
PDB header: oxidoreductaseChain: B: PDB Molecule: dihydrofolate reductase;PDBTitle: crystal structure of streptococcus pneumoniae dihydrofolate2 reductase - sp9 mutant
81 c1cliD_
not modelled
11.8
32
PDB header: ligaseChain: D: PDB Molecule: protein (phosphoribosyl-aminoimidazole synthetase);PDBTitle: x-ray crystal structure of aminoimidazole ribonucleotide synthetase2 (purm), from the e. coli purine biosynthetic pathway, at 2.5 a3 resolution
82 d1jcea2
not modelled
11.5
29
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP7083 d1fe0a_
not modelled
11.5
21
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain84 d1xpja_
not modelled
11.4
8
Fold: HAD-likeSuperfamily: HAD-likeFamily: Hypothetical protein VC023285 d1xbta1
not modelled
11.4
10
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Type II thymidine kinase86 c2pr7A_
not modelled
11.3
11
PDB header: hydrolaseChain: A: PDB Molecule: haloacid dehalogenase/epoxide hydrolase family;PDBTitle: crystal structure of uncharacterized protein (np_599989.1) from2 corynebacterium glutamicum atcc 13032 kitasato at 1.44 a resolution
87 d2b7oa1
not modelled
11.2
46
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class-II DAHP synthetase88 c3u1nC_
not modelled
11.0
10
PDB header: hydrolaseChain: C: PDB Molecule: sam domain and hd domain-containing protein 1;PDBTitle: structure of the catalytic core of human samhd1
89 c2jtqA_
not modelled
11.0
22
PDB header: transferaseChain: A: PDB Molecule: phage shock protein e;PDBTitle: rhodanese from e.coli
90 c3fozB_
not modelled
11.0
27
PDB header: transferase/rnaChain: B: PDB Molecule: trna delta(2)-isopentenylpyrophosphate transferase;PDBTitle: structure of e. coli isopentenyl-trna transferase in complex with e.2 coli trna(phe)
91 c3fryB_
not modelled
11.0
35
PDB header: hydrolaseChain: B: PDB Molecule: probable copper-exporting p-type atpase a;PDBTitle: crystal structure of the copa c-terminal metal binding domain
92 c2i7dB_
not modelled
10.9
15
PDB header: hydrolaseChain: B: PDB Molecule: 5'(3')-deoxyribonucleotidase, cytosolic type;PDBTitle: structure of human cytosolic deoxyribonucleotidase in2 complex with deoxyuridine, alf4 and mg2+
93 d1z5ga1
not modelled
10.8
21
Fold: HAD-likeSuperfamily: HAD-likeFamily: Class B acid phosphatase, AphA94 c2kmvA_
not modelled
10.7
21
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the nucleotide binding domain of the2 human menkes protein in the atp-free form
95 c2v9yA_
not modelled
10.7
23
PDB header: ligaseChain: A: PDB Molecule: phosphoribosylformylglycinamidine cyclo-ligase;PDBTitle: human aminoimidazole ribonucleotide synthetase
96 c2z01A_
not modelled
10.7
27
PDB header: ligaseChain: A: PDB Molecule: phosphoribosylformylglycinamidine cyclo-ligase;PDBTitle: crystal structure of phosphoribosylaminoimidazole2 synthetase from geobacillus kaustophilus
97 d1nlna_
not modelled
10.5
13
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Adenain-like98 d2bsza1
not modelled
10.4
27
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Arylamine N-acetyltransferase99 d1u6za1
not modelled
10.3
10
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: Ppx associated domain