| 1 |
|
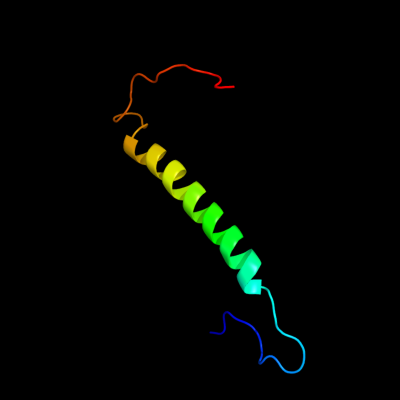
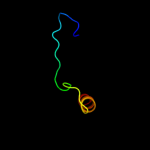
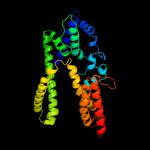
PDB 1qle chain B
Region: 322 - 376
Aligned: 55
Modelled: 55
Confidence: 36.5%
Identity: 16%
PDB header:oxidoreductase/immune system
Chain: B: PDB Molecule:cytochrome c oxidase polypeptide ii;
PDBTitle: cryo-structure of the paracoccus denitrificans four-subunit2 cytochrome c oxidase in the completely oxidized state3 complexed with an antibody fv fragment
Phyre2
| 2 |
|
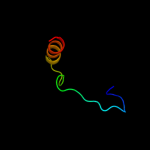
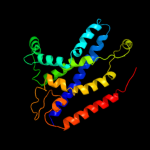
PDB 1ar1 chain B
Region: 322 - 376
Aligned: 55
Modelled: 55
Confidence: 36.5%
Identity: 16%
PDB header:complex (oxidoreductase/antibody)
Chain: B: PDB Molecule:cytochrome c oxidase;
PDBTitle: structure at 2.7 angstrom resolution of the paracoccus2 denitrificans two-subunit cytochrome c oxidase complexed3 with an antibody fv fragment
Phyre2
| 3 |
|
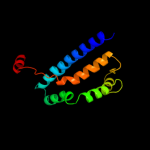
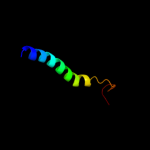
PDB 3m7b chain A
Region: 129 - 370
Aligned: 188
Modelled: 197
Confidence: 17.6%
Identity: 11%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:tellurite resistance protein teha homolog;
PDBTitle: crystal structure of plant slac1 homolog teha
Phyre2
| 4 |
|
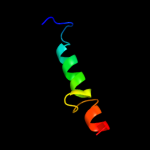
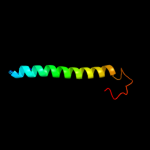
PDB 1ejx chain C domain 2
Region: 307 - 338
Aligned: 32
Modelled: 32
Confidence: 16.1%
Identity: 25%
Fold: TIM beta/alpha-barrel
Superfamily: Metallo-dependent hydrolases
Family: alpha-subunit of urease, catalytic domain
Phyre2
| 5 |
|
PDB 1e9y chain B domain 2
Region: 307 - 338
Aligned: 32
Modelled: 32
Confidence: 15.3%
Identity: 16%
Fold: TIM beta/alpha-barrel
Superfamily: Metallo-dependent hydrolases
Family: alpha-subunit of urease, catalytic domain
Phyre2
| 6 |
|
PDB 2r6g chain F domain 2
Region: 76 - 201
Aligned: 125
Modelled: 126
Confidence: 12.6%
Identity: 10%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 7 |
|
PDB 2hku chain A domain 2
Region: 106 - 139
Aligned: 28
Modelled: 34
Confidence: 11.7%
Identity: 25%
Fold: Tetracyclin repressor-like, C-terminal domain
Superfamily: Tetracyclin repressor-like, C-terminal domain
Family: Tetracyclin repressor-like, C-terminal domain
Phyre2
| 8 |
|
PDB 2xq2 chain A
Region: 3 - 375
Aligned: 295
Modelled: 304
Confidence: 10.7%
Identity: 13%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
Phyre2
| 9 |
|
PDB 2kta chain A
Region: 40 - 58
Aligned: 19
Modelled: 19
Confidence: 8.9%
Identity: 21%
PDB header:hydrolase
Chain: A: PDB Molecule:putative helicase;
PDBTitle: solution nmr structure of a domain of protein a6ky75 from bacteroides2 vulgatus, northeast structural genomics target bvr106a
Phyre2
| 10 |
|
PDB 2f3o chain B
Region: 42 - 62
Aligned: 21
Modelled: 21
Confidence: 8.0%
Identity: 5%
PDB header:unknown function
Chain: B: PDB Molecule:pyruvate formate-lyase 2;
PDBTitle: crystal structure of a glycyl radical enzyme from archaeoglobus2 fulgidus
Phyre2
| 11 |
|
PDB 3fh6 chain F
Region: 75 - 287
Aligned: 212
Modelled: 213
Confidence: 7.9%
Identity: 9%
PDB header:transport protein
Chain: F: PDB Molecule:maltose transport system permease protein malf;
PDBTitle: crystal structure of the resting state maltose transporter from e.2 coli
Phyre2
| 12 |
|
PDB 2knc chain A
Region: 337 - 376
Aligned: 40
Modelled: 40
Confidence: 7.3%
Identity: 15%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 13 |
|
PDB 3dtu chain B domain 2
Region: 321 - 376
Aligned: 56
Modelled: 56
Confidence: 6.7%
Identity: 20%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 14 |
|
PDB 2iub chain A domain 2
Region: 333 - 361
Aligned: 29
Modelled: 29
Confidence: 6.2%
Identity: 17%
Fold: Transmembrane helix hairpin
Superfamily: Magnesium transport protein CorA, transmembrane region
Family: Magnesium transport protein CorA, transmembrane region
Phyre2
| 15 |
|
PDB 2v5i chain A
Region: 52 - 59
Aligned: 8
Modelled: 8
Confidence: 5.9%
Identity: 25%
PDB header:viral protein
Chain: A: PDB Molecule:salmonella typhimurium db7155 bacteriophage det7
PDBTitle: structure of the receptor-binding protein of bacteriophage2 det7: a podoviral tailspike in a myovirus
Phyre2
| 16 |
|
PDB 3hfx chain A
Region: 32 - 193
Aligned: 135
Modelled: 148
Confidence: 5.6%
Identity: 13%
PDB header:transport protein
Chain: A: PDB Molecule:l-carnitine/gamma-butyrobetaine antiporter;
PDBTitle: crystal structure of carnitine transporter
Phyre2