1 d1dj0a_
100.0
100
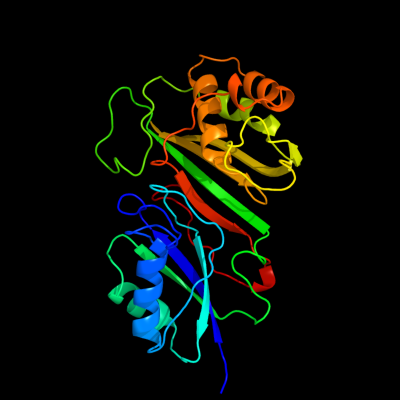
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase I TruA2 c1vs3B_
100.0
34
PDB header: isomeraseChain: B: PDB Molecule: trna pseudouridine synthase a;PDBTitle: crystal structure of the trna pseudouridine synthase trua from thermus2 thermophilus hb8
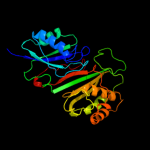
3 c2v9kA_
91.6
17
PDB header: lyaseChain: A: PDB Molecule: uncharacterized protein flj32312;PDBTitle: crystal structure of human pus10, a novel pseudouridine2 synthase.
4 c1qysA_
54.5
15
PDB header: de novo proteinChain: A: PDB Molecule: top7;PDBTitle: crystal structure of top7: a computationally designed2 protein with a novel fold
5 c2jvfA_
49.5
22
PDB header: de novo proteinChain: A: PDB Molecule: de novo protein m7;PDBTitle: solution structure of m7, a computationally-designed2 artificial protein
6 d2hh8a1
41.2
12
Fold: YdfO-likeSuperfamily: YdfO-likeFamily: YdfO-like7 d1q9ja1
21.9
15
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: NRPS condensation domain (amide synthase)8 c2kruA_
20.7
19
PDB header: oxidoreductaseChain: A: PDB Molecule: light-independent protochlorophyllide reductasePDBTitle: solution nmr structure of the pcp_red domain of light-2 independent protochlorophyllide reductase subunit b from3 chlorobium tepidum. northeast structural genomics4 consortium target ctr69a (casp target)
9 d1e8ya3
19.1
13
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ras-binding domain, RBD10 d1e88a3
18.0
40
Fold: FnI-like domainSuperfamily: FnI-like domainFamily: Fibronectin type I module11 d1nija2
17.3
11
Fold: Hypothetical protein YjiA, C-terminal domainSuperfamily: Hypothetical protein YjiA, C-terminal domainFamily: Hypothetical protein YjiA, C-terminal domain12 d1l5aa1
13.3
28
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: NRPS condensation domain (amide synthase)13 c2l09A_
12.4
3
PDB header: oxidoreductaseChain: A: PDB Molecule: asr4154 protein;PDBTitle: solution nmr structure of protein asr4154 from nostoc sp. pcc71202 northeast structural genomics consortium target id nsr143
14 c1l5aA_
12.1
19
PDB header: biosynthetic proteinChain: A: PDB Molecule: amide synthase;PDBTitle: crystal structure of vibh, an nrps condensation enzyme
15 d1u02a_
12.1
20
Fold: HAD-likeSuperfamily: HAD-likeFamily: Trehalose-phosphatase16 d1y6va1
10.7
16
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Alkaline phosphatase17 c2xhgA_
9.9
14
PDB header: isomeraseChain: A: PDB Molecule: tyrocidine synthetase a;PDBTitle: crystal structure of the epimerization domain from the initiation2 module of tyrocidine biosynthesis
18 d2gjva1
9.3
33
Fold: Phage tail protein-likeSuperfamily: Phage tail protein-likeFamily: STM4215-like19 c2gjvF_
9.3
33
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: putative cytoplasmic protein;PDBTitle: crystal structure of a protein of unknown function from salmonella2 typhimurium
20 c1q9jA_
7.7
15
PDB header: ligaseChain: A: PDB Molecule: polyketide synthase associated protein 5;PDBTitle: structure of polyketide synthase associated protein 5 from2 mycobacterium tuberculosis
21 c3ni2A_
not modelled
7.5
7
PDB header: ligaseChain: A: PDB Molecule: 4-coumarate:coa ligase;PDBTitle: crystal structures and enzymatic mechanisms of a populus tomentosa 4-2 coumarate:coa ligase
22 c1e88A_
not modelled
7.5
40
PDB header: cell adhesionChain: A: PDB Molecule: fibronectin;PDBTitle: solution structure of 6f11f22f2, a compact three-module2 fragment of the gelatin-binding domain of human fibronectin
23 d2fbla1
not modelled
7.4
21
Fold: CYTH-like phosphatasesSuperfamily: CYTH-like phosphatasesFamily: CYTH domain24 c2wwaJ_
not modelled
7.4
19
PDB header: ribosomeChain: J: PDB Molecule: 60s ribosomal protein l19;PDBTitle: cryo-em structure of idle yeast ssh1 complex bound to the2 yeast 80s ribosome
25 c2omlA_
not modelled
7.0
20
PDB header: isomeraseChain: A: PDB Molecule: ribosomal large subunit pseudouridine synthase e;PDBTitle: crystal structure of e. coli pseudouridine synthase rlue
26 c2jv4A_
not modelled
6.8
15
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis/trans isomerase;PDBTitle: structure characterisation of pina ww domain and comparison2 with other group iv ww domains, pin1 and ess1
27 d1xb2b2
not modelled
6.7
16
Fold: EF-Ts domain-likeSuperfamily: Elongation factor Ts (EF-Ts), dimerisation domainFamily: Elongation factor Ts (EF-Ts), dimerisation domain28 d1qlma_
not modelled
6.3
31
Fold: Methenyltetrahydromethanopterin cyclohydrolaseSuperfamily: Methenyltetrahydromethanopterin cyclohydrolaseFamily: Methenyltetrahydromethanopterin cyclohydrolase29 c3dh3C_
not modelled
6.3
10
PDB header: isomerase/rnaChain: C: PDB Molecule: ribosomal large subunit pseudouridine synthase f;PDBTitle: crystal structure of rluf in complex with a 22 nucleotide2 rna substrate
30 d2csha2
not modelled
6.1
20
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H231 c1kskA_
not modelled
6.1
20
PDB header: lyaseChain: A: PDB Molecule: ribosomal small subunit pseudouridine synthase a;PDBTitle: structure of rsua
32 d1owla1
not modelled
5.9
33
Fold: Cryptochrome/photolyase FAD-binding domainSuperfamily: Cryptochrome/photolyase FAD-binding domainFamily: Cryptochrome/photolyase FAD-binding domain33 d1dnpa1
not modelled
5.7
27
Fold: Cryptochrome/photolyase FAD-binding domainSuperfamily: Cryptochrome/photolyase FAD-binding domainFamily: Cryptochrome/photolyase FAD-binding domain34 c3l9aX_
not modelled
5.6
32
PDB header: structural genomics, unknown functionChain: X: PDB Molecule: uncharacterized protein;PDBTitle: structure of the c-terminal domain from a streptococcus2 mutans hypothetical
35 d1bdga2
not modelled
5.5
14
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase36 d1czan2
not modelled
5.4
17
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase37 c1unyA_
not modelled
5.3
42
PDB header: four helix bundleChain: A: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
38 d1iwga1
not modelled
5.3
14
Fold: Ferredoxin-likeSuperfamily: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomainsFamily: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomains39 c2cooA_
not modelled
5.2
10
PDB header: transferaseChain: A: PDB Molecule: lipoamide acyltransferase component of branched-PDBTitle: solution structure of the e3_binding domain of2 dihydrolipoamide branched chaintransacylase
40 c3a52A_
not modelled
5.2
11
PDB header: hydrolaseChain: A: PDB Molecule: cold-active alkaline phosphatase;PDBTitle: crystal structure of cold-active alkailne phosphatase from2 psychrophile shewanella sp.
41 c3umvB_
not modelled
5.1
13
PDB header: lyaseChain: B: PDB Molecule: deoxyribodipyrimidine photo-lyase;PDBTitle: eukaryotic class ii cpd photolyase structure reveals a basis for2 improved uv-tolerance in plants