| 1 |
|
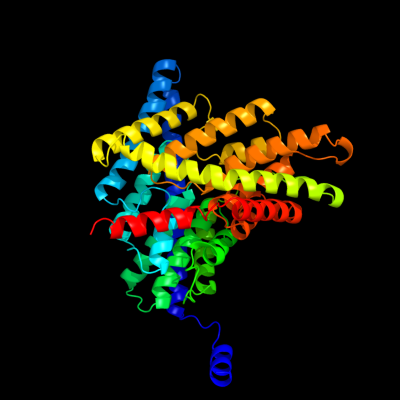
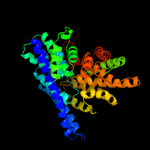
PDB 1ots chain A
Region: 17 - 460
Aligned: 444
Modelled: 444
Confidence: 100.0%
Identity: 100%
Fold: Clc chloride channel
Superfamily: Clc chloride channel
Family: Clc chloride channel
Phyre2
| 2 |
|
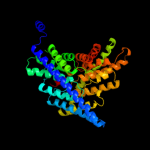
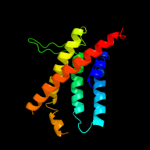
PDB 2ht2 chain B
Region: 18 - 458
Aligned: 441
Modelled: 441
Confidence: 100.0%
Identity: 100%
PDB header:membrane protein
Chain: B: PDB Molecule:h(+)/cl(-) exchange transporter clca;
PDBTitle: structure of the escherichia coli clc chloride channel2 y445h mutant and fab complex
Phyre2
| 3 |
|
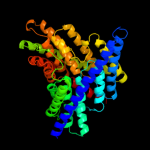
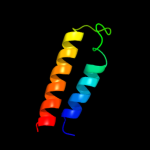
PDB 1kpl chain A
Region: 31 - 460
Aligned: 430
Modelled: 430
Confidence: 100.0%
Identity: 81%
Fold: Clc chloride channel
Superfamily: Clc chloride channel
Family: Clc chloride channel
Phyre2
| 4 |
|
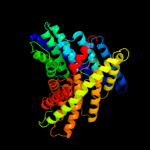
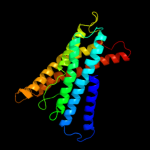
PDB 3nd0 chain A
Region: 32 - 458
Aligned: 423
Modelled: 427
Confidence: 100.0%
Identity: 40%
PDB header:transport protein
Chain: A: PDB Molecule:sll0855 protein;
PDBTitle: x-ray crystal structure of a slow cyanobacterial cl-/h+ antiporter
Phyre2
| 5 |
|
PDB 3org chain B
Region: 33 - 467
Aligned: 403
Modelled: 421
Confidence: 100.0%
Identity: 23%
PDB header:transport protein
Chain: B: PDB Molecule:cmclc;
PDBTitle: crystal structure of a eukaryotic clc transporter
Phyre2
| 6 |
|
PDB 3p5n chain A
Region: 133 - 278
Aligned: 131
Modelled: 146
Confidence: 26.9%
Identity: 21%
PDB header:transport protein
Chain: A: PDB Molecule:riboflavin uptake protein;
PDBTitle: structure and mechanism of the s component of a bacterial ecf2 transporter
Phyre2
| 7 |
|
PDB 2ksf chain A
Region: 217 - 267
Aligned: 50
Modelled: 51
Confidence: 21.4%
Identity: 12%
PDB header:transferase
Chain: A: PDB Molecule:sensor protein kdpd;
PDBTitle: backbone structure of the membrane domain of e. coli2 histidine kinase receptor kdpd, center for structures of3 membrane proteins (csmp) target 4312c
Phyre2
| 8 |
|
PDB 1v54 chain A
Region: 212 - 451
Aligned: 240
Modelled: 240
Confidence: 12.9%
Identity: 13%
Fold: Cytochrome c oxidase subunit I-like
Superfamily: Cytochrome c oxidase subunit I-like
Family: Cytochrome c oxidase subunit I-like
Phyre2
| 9 |
|
PDB 3c4r chain C
Region: 419 - 466
Aligned: 48
Modelled: 48
Confidence: 10.9%
Identity: 19%
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of an uncharacterized protein encoded by2 cryptic prophage
Phyre2
| 10 |
|
PDB 2dw3 chain A
Region: 349 - 363
Aligned: 15
Modelled: 15
Confidence: 7.7%
Identity: 40%
PDB header:photosynthesis
Chain: A: PDB Molecule:intrinsic membrane protein pufx;
PDBTitle: solution structure of the rhodobacter sphaeroides pufx2 membrane protein
Phyre2
| 11 |
|
PDB 3mk7 chain F
Region: 33 - 96
Aligned: 64
Modelled: 64
Confidence: 7.5%
Identity: 17%
PDB header:oxidoreductase
Chain: F: PDB Molecule:cytochrome c oxidase, cbb3-type, subunit p;
PDBTitle: the structure of cbb3 cytochrome oxidase
Phyre2
| 12 |
|
PDB 3din chain D
Region: 11 - 64
Aligned: 53
Modelled: 54
Confidence: 5.9%
Identity: 11%
PDB header:membrane protein, protein transport
Chain: D: PDB Molecule:preprotein translocase subunit sece;
PDBTitle: crystal structure of the protein-translocation complex formed by the2 secy channel and the seca atpase
Phyre2
| 13 |
|
PDB 2akh chain Z
Region: 7 - 65
Aligned: 58
Modelled: 59
Confidence: 5.6%
Identity: 12%
PDB header:protein transport
Chain: Z: PDB Molecule:preprotein translocase sece subunit;
PDBTitle: normal mode-based flexible fitted coordinates of a non-2 translocating secyeg protein-conducting channel into the3 cryo-em map of a secyeg-nascent chain-70s ribosome complex4 from e. coli
Phyre2
| 14 |
|
PDB 2kjf chain A
Region: 133 - 161
Aligned: 29
Modelled: 29
Confidence: 5.3%
Identity: 21%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:carnocyclin-a;
PDBTitle: the solution structure of the circular bacteriocin2 carnocyclin a (ccla)
Phyre2
| 15 |
|
PDB 2yl4 chain A
Region: 20 - 100
Aligned: 81
Modelled: 81
Confidence: 5.1%
Identity: 9%
PDB header:membrane protein
Chain: A: PDB Molecule:atp-binding cassette sub-family b member 10,
PDBTitle: structure of the human mitochondrial abc transporter, abcb10
Phyre2