| 1 |
|
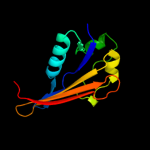
PDB 3ly8 chain A
Region: 110 - 211
Aligned: 84
Modelled: 85
Confidence: 99.5%
Identity: 8%
PDB header:signaling protein
Chain: A: PDB Molecule:transcriptional activator cadc;
PDBTitle: crystal structure of mutant d471e of the periplasmic domain of cadc
Phyre2
| 2 |
|
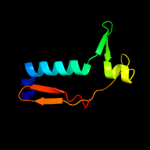
PDB 2hqs chain A domain 2
Region: 100 - 213
Aligned: 99
Modelled: 114
Confidence: 99.2%
Identity: 16%
Fold: Anticodon-binding domain-like
Superfamily: TolB, N-terminal domain
Family: TolB, N-terminal domain
Phyre2
| 3 |
|
PDB 2w8b chain B
Region: 103 - 213
Aligned: 100
Modelled: 111
Confidence: 87.8%
Identity: 19%
PDB header:protein transport/membrane protein
Chain: B: PDB Molecule:protein tolb;
PDBTitle: crystal structure of processed tolb in complex with pal
Phyre2
| 4 |
|
PDB 2ijr chain A domain 1
Region: 6 - 37
Aligned: 32
Modelled: 32
Confidence: 85.2%
Identity: 38%
Fold: Api92-like
Superfamily: Api92-like
Family: Api92-like
Phyre2
| 5 |
|
PDB 2ifs chain A
Region: 192 - 207
Aligned: 16
Modelled: 16
Confidence: 35.2%
Identity: 13%
PDB header:signaling protein
Chain: A: PDB Molecule:wiskott-aldrich syndrome protien ineracting
PDBTitle: structure of the n-wasp evh1 domain in complex with an2 extended wip peptide
Phyre2
| 6 |
|
PDB 2iqi chain A domain 1
Region: 118 - 211
Aligned: 63
Modelled: 63
Confidence: 24.2%
Identity: 19%
Fold: Anticodon-binding domain-like
Superfamily: XCC0632-like
Family: XCC0632-like
Phyre2
| 7 |
|
PDB 1mke chain A domain 1
Region: 190 - 207
Aligned: 18
Modelled: 18
Confidence: 17.1%
Identity: 11%
Fold: PH domain-like barrel
Superfamily: PH domain-like
Family: Enabled/VASP homology 1 domain (EVH1 domain)
Phyre2
| 8 |
|
PDB 1flg chain A
Region: 191 - 205
Aligned: 15
Modelled: 15
Confidence: 13.0%
Identity: 13%
Fold: 8-bladed beta-propeller
Superfamily: Quinoprotein alcohol dehydrogenase-like
Family: Quinoprotein alcohol dehydrogenase-like
Phyre2
| 9 |
|
PDB 3gr1 chain A
Region: 96 - 194
Aligned: 91
Modelled: 99
Confidence: 11.8%
Identity: 19%
PDB header:membrane protein
Chain: A: PDB Molecule:protein prgh;
PDBTitle: periplamic domain of the t3ss inner membrane protein prgh2 from s.typhimurium (fragment 170-392)
Phyre2
| 10 |
|
PDB 2ivz chain D
Region: 98 - 213
Aligned: 101
Modelled: 116
Confidence: 9.3%
Identity: 18%
PDB header:protein transport/hydrolase
Chain: D: PDB Molecule:protein tolb;
PDBTitle: structure of tolb in complex with a peptide of the colicin2 e9 t-domain
Phyre2
| 11 |
|
PDB 2be1 chain A
Region: 191 - 205
Aligned: 15
Modelled: 15
Confidence: 8.0%
Identity: 33%
PDB header:transcription
Chain: A: PDB Molecule:serine/threonine-protein kinase/endoribonuclease ire1;
PDBTitle: structure of the compact lumenal domain of yeast ire1
Phyre2
| 12 |
|
PDB 1kv9 chain A domain 2
Region: 191 - 205
Aligned: 15
Modelled: 15
Confidence: 8.0%
Identity: 33%
Fold: 8-bladed beta-propeller
Superfamily: Quinoprotein alcohol dehydrogenase-like
Family: Quinoprotein alcohol dehydrogenase-like
Phyre2
| 13 |
|
PDB 3tek chain A
Region: 123 - 209
Aligned: 66
Modelled: 75
Confidence: 7.6%
Identity: 18%
PDB header:dna binding protein
Chain: A: PDB Molecule:thermodbp-single stranded dna binding protein;
PDBTitle: thermodbp: a non-canonical single-stranded dna binding protein with a2 novel structure and mechanism
Phyre2
| 14 |
|
PDB 2ad6 chain A domain 1
Region: 191 - 205
Aligned: 15
Modelled: 15
Confidence: 7.1%
Identity: 13%
Fold: 8-bladed beta-propeller
Superfamily: Quinoprotein alcohol dehydrogenase-like
Family: Quinoprotein alcohol dehydrogenase-like
Phyre2
| 15 |
|
PDB 3d33 chain B
Region: 190 - 206
Aligned: 17
Modelled: 16
Confidence: 7.1%
Identity: 6%
PDB header:unknown function
Chain: B: PDB Molecule:domain of unknown function with an immunoglobulin-like
PDBTitle: crystal structure of a duf3244 family protein with an immunoglobulin-2 like beta-sandwich fold (bvu_0276) from bacteroides vulgatus atcc3 8482 at 1.70 a resolution
Phyre2
| 16 |
|
PDB 1kv9 chain A
Region: 191 - 205
Aligned: 15
Modelled: 15
Confidence: 7.0%
Identity: 33%
PDB header:oxidoreductase
Chain: A: PDB Molecule:type ii quinohemoprotein alcohol dehydrogenase;
PDBTitle: structure at 1.9 a resolution of a quinohemoprotein alcohol2 dehydrogenase from pseudomonas putida hk5
Phyre2
| 17 |
|
PDB 1bml chain C domain 1
Region: 105 - 182
Aligned: 75
Modelled: 78
Confidence: 6.4%
Identity: 9%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: Staphylokinase/streptokinase
Family: Staphylokinase/streptokinase
Phyre2
| 18 |
|
PDB 3o47 chain A
Region: 84 - 139
Aligned: 55
Modelled: 56
Confidence: 6.0%
Identity: 18%
PDB header:hydrolase, hydrolase activator
Chain: A: PDB Molecule:adp-ribosylation factor gtpase-activating protein 1, adp-
PDBTitle: crystal structure of arfgap1-arf1 fusion protein
Phyre2
| 19 |
|
PDB 1w6s chain A
Region: 191 - 205
Aligned: 15
Modelled: 15
Confidence: 5.9%
Identity: 20%
Fold: 8-bladed beta-propeller
Superfamily: Quinoprotein alcohol dehydrogenase-like
Family: Quinoprotein alcohol dehydrogenase-like
Phyre2
| 20 |
|
PDB 3sd2 chain A
Region: 190 - 206
Aligned: 17
Modelled: 17
Confidence: 5.4%
Identity: 29%
PDB header:unknown function
Chain: A: PDB Molecule:putative member of duf3244 protein family;
PDBTitle: crystal structure of a putative member of duf3244 protein family2 (bt_3571) from bacteroides thetaiotaomicron vpi-5482 at 1.40 a3 resolution
Phyre2
| 21 |
|