| 1 |
|
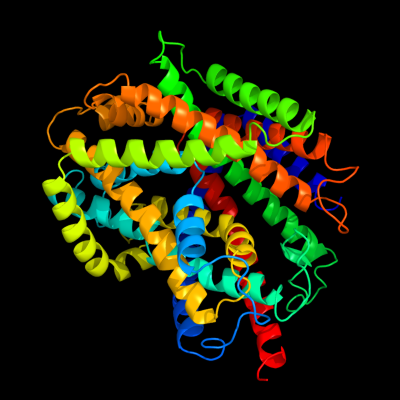
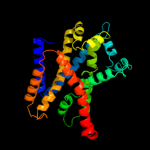
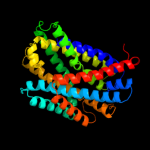
PDB 3hfx chain A
Region: 12 - 504
Aligned: 493
Modelled: 493
Confidence: 100.0%
Identity: 100%
PDB header:transport protein
Chain: A: PDB Molecule:l-carnitine/gamma-butyrobetaine antiporter;
PDBTitle: crystal structure of carnitine transporter
Phyre2
| 2 |
|
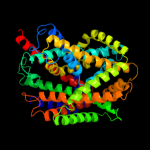
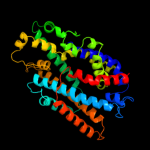
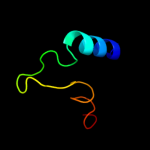
PDB 2w8a chain C
Region: 9 - 504
Aligned: 484
Modelled: 495
Confidence: 100.0%
Identity: 26%
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
Phyre2
| 3 |
|
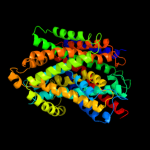
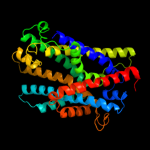
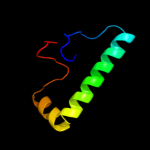
PDB 2xq2 chain A
Region: 141 - 492
Aligned: 336
Modelled: 352
Confidence: 98.2%
Identity: 14%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
Phyre2
| 4 |
|
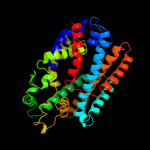
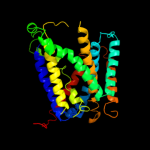
PDB 2a65 chain A domain 1
Region: 224 - 504
Aligned: 266
Modelled: 281
Confidence: 97.6%
Identity: 15%
Fold: SNF-like
Superfamily: SNF-like
Family: SNF-like
Phyre2
| 5 |
|
PDB 3dh4 chain A
Region: 142 - 492
Aligned: 339
Modelled: 351
Confidence: 97.3%
Identity: 13%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
Phyre2
| 6 |
|
PDB 2jln chain A
Region: 140 - 503
Aligned: 333
Modelled: 343
Confidence: 97.1%
Identity: 9%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
| 7 |
|
PDB 3lrc chain C
Region: 142 - 504
Aligned: 313
Modelled: 313
Confidence: 85.1%
Identity: 12%
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
Phyre2
| 8 |
|
PDB 3gia chain A
Region: 142 - 504
Aligned: 331
Modelled: 331
Confidence: 83.3%
Identity: 9%
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized protein mj0609;
PDBTitle: crystal structure of apct transporter
Phyre2
| 9 |
|
PDB 3gzt chain F
Region: 311 - 320
Aligned: 10
Modelled: 10
Confidence: 23.6%
Identity: 40%
PDB header:virus
Chain: F: PDB Molecule:outer capsid glycoprotein vp7;
PDBTitle: vp7 recoated rotavirus dlp
Phyre2
| 10 |
|
PDB 2kvl chain A
Region: 311 - 320
Aligned: 10
Modelled: 10
Confidence: 23.3%
Identity: 40%
PDB header:viral protein
Chain: A: PDB Molecule:major outer capsid protein vp7;
PDBTitle: nmr structure of the c-terminal domain of vp7
Phyre2
| 11 |
|
PDB 1zza chain A
Region: 148 - 188
Aligned: 40
Modelled: 41
Confidence: 11.4%
Identity: 20%
PDB header:membrane protein
Chain: A: PDB Molecule:stannin;
PDBTitle: solution nmr structure of the membrane protein stannin
Phyre2
| 12 |
|
PDB 1qmv chain A
Region: 75 - 87
Aligned: 13
Modelled: 13
Confidence: 6.6%
Identity: 31%
Fold: Thioredoxin fold
Superfamily: Thioredoxin-like
Family: Glutathione peroxidase-like
Phyre2
| 13 |
|
PDB 1xb4 chain C
Region: 115 - 186
Aligned: 56
Modelled: 57
Confidence: 6.5%
Identity: 20%
PDB header:unknown function
Chain: C: PDB Molecule:hypothetical 23.6 kda protein in yuh1-ura8
PDBTitle: crystal structure of subunit vps25 of the endosomal2 trafficking complex escrt-ii
Phyre2
| 14 |
|
PDB 3f1i chain H
Region: 331 - 344
Aligned: 14
Modelled: 14
Confidence: 5.9%
Identity: 36%
PDB header:protein binding
Chain: H: PDB Molecule:hepatocyte growth factor-regulated tyrosine kinase
PDBTitle: human escrt-0 core complex
Phyre2
| 15 |
|
PDB 1qq2 chain A
Region: 75 - 87
Aligned: 13
Modelled: 13
Confidence: 5.9%
Identity: 23%
Fold: Thioredoxin fold
Superfamily: Thioredoxin-like
Family: Glutathione peroxidase-like
Phyre2
| 16 |
|
PDB 3eyp chain B
Region: 120 - 147
Aligned: 28
Modelled: 28
Confidence: 5.7%
Identity: 25%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative alpha-l-fucosidase;
PDBTitle: crystal structure of putative alpha-l-fucosidase from bacteroides2 thetaiotaomicron
Phyre2
| 17 |
|
PDB 1jxh chain A
Region: 330 - 346
Aligned: 17
Modelled: 17
Confidence: 5.6%
Identity: 24%
Fold: Ribokinase-like
Superfamily: Ribokinase-like
Family: Thiamin biosynthesis kinases
Phyre2