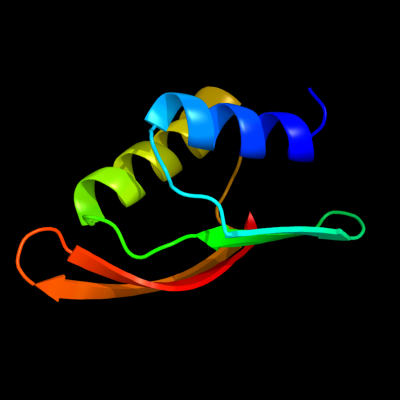
| 1 | c1uv7A_
|
|
 |
99.8 |
11 |
PDB header:transport
Chain: A: PDB Molecule:general secretion pathway protein m;
PDBTitle: periplasmic domain of epsm from vibrio cholerae
|
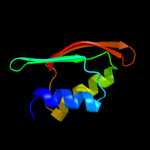
| 2 | d1uv7a_
|
|
 |
99.8 |
11 |
Fold:RRF/tRNA synthetase additional domain-like
Superfamily:General secretion pathway protein M, EpsM
Family:General secretion pathway protein M, EpsM |
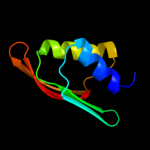
| 3 | c2rjzA_
|
|
 |
97.7 |
9 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:pilo protein;
PDBTitle: crystal structure of the type 4 fimbrial biogenesis protein pilo from2 pseudomonas aeruginosa
|
| 4 | c2rddB_
|
|
 |
57.7 |
17 |
PDB header:membrane protein/transport protein
Chain: B: PDB Molecule:upf0092 membrane protein yajc;
PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
|
| 5 | c2y9kG_
|
|
 |
47.2 |
9 |
PDB header:protein transport
Chain: G: PDB Molecule:protein invg;
PDBTitle: three-dimensional model of salmonella's needle complex at2 subnanometer resolution
|
| 6 | d1bjna_
|
|
 |
29.5 |
9 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 7 | c3gr5A_
|
|
 |
28.1 |
10 |
PDB header:membrane protein
Chain: A: PDB Molecule:escc;
PDBTitle: periplasmic domain of the outer membrane secretin escc from2 enteropathogenic e.coli (epec)
|
| 8 | d1ppjc2
|
|
 |
26.1 |
9 |
Fold:Heme-binding four-helical bundle
Superfamily:Transmembrane di-heme cytochromes
Family:Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase) |
| 9 | d1tp6a_
|
|
 |
24.9 |
22 |
Fold:Cystatin-like
Superfamily:NTF2-like
Family:PA1314-like |
| 10 | d1dnla_
|
|
 |
24.9 |
15 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:PNP-oxidase like |
| 11 | c3hd7A_
|
|
 |
22.9 |
22 |
PDB header:exocytosis
Chain: A: PDB Molecule:vesicle-associated membrane protein 2;
PDBTitle: helical extension of the neuronal snare complex into the membrane,2 spacegroup c 1 2 1
|
| 12 | d1yioa1
|
|
 |
21.4 |
10 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:C-terminal effector domain of the bipartite response regulators
Family:GerE-like (LuxR/UhpA family of transcriptional regulators) |
| 13 | d2c0ra1
|
|
 |
20.2 |
7 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 14 | d1x4pa1
|
|
 |
18.6 |
18 |
Fold:Surp module (SWAP domain)
Superfamily:Surp module (SWAP domain)
Family:Surp module (SWAP domain) |
| 15 | d1nrga_
|
|
 |
18.1 |
11 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:PNP-oxidase like |
| 16 | c1nrgA_
|
|
 |
18.1 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyridoxine 5'-phosphate oxidase;
PDBTitle: structure and properties of recombinant human pyridoxine-5'-phosphate2 oxidase
|
| 17 | d1t9ma_
|
|
 |
17.1 |
17 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:PNP-oxidase like |
| 18 | c2kncA_
|
|
 |
16.5 |
25 |
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
|
| 19 | d1hywa_
|
|
 |
14.4 |
5 |
Fold:gpW/XkdW-like
Superfamily:Head-to-tail joining protein W, gpW
Family:Head-to-tail joining protein W, gpW |
| 20 | d1bccc3
|
|
 |
12.8 |
11 |
Fold:Heme-binding four-helical bundle
Superfamily:Transmembrane di-heme cytochromes
Family:Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase) |
| 21 | d1v54d_ |
|
not modelled |
12.5 |
27 |
Fold:Single transmembrane helix
Superfamily:Mitochondrial cytochrome c oxidase subunit IV
Family:Mitochondrial cytochrome c oxidase subunit IV |
| 22 | c3mn7S_ |
|
not modelled |
12.1 |
20 |
PDB header:contractile protein/protein binding
Chain: S: PDB Molecule:spire ddd;
PDBTitle: structures of actin-bound wh2 domains of spire and the implication for2 filament nucleation
|
| 23 | c2rnjA_ |
|
not modelled |
11.8 |
19 |
PDB header:transcription
Chain: A: PDB Molecule:response regulator protein vrar;
PDBTitle: nmr structure of the s. aureus vrar dna binding domain
|
| 24 | c3mn5S_ |
|
not modelled |
11.8 |
20 |
PDB header:contractile protein/protein binding
Chain: S: PDB Molecule:protein spire;
PDBTitle: structures of actin-bound wh2 domains of spire and the implication for2 filament nucleation
|
| 25 | c3qm2A_ |
|
not modelled |
11.4 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoserine aminotransferase;
PDBTitle: 2.25 angstrom crystal structure of phosphoserine aminotransferase2 (serc) from salmonella enterica subsp. enterica serovar typhimurium
|
| 26 | c2d89A_ |
|
not modelled |
10.7 |
19 |
PDB header:structural protein, protein binding
Chain: A: PDB Molecule:ehbp1 protein;
PDBTitle: solution structure of the ch domain from human eh domain2 binding protein 1
|
| 27 | c2y69Q_ |
|
not modelled |
10.6 |
27 |
PDB header:electron transport
Chain: Q: PDB Molecule:cytochrome c oxidase subunit 4 isoform 1;
PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
|
| 28 | c2w7vB_ |
|
not modelled |
10.5 |
11 |
PDB header:transport protein
Chain: B: PDB Molecule:general secretion pathway protein l;
PDBTitle: periplasmic domain of epsl from vibrio parahaemolyticus
|
| 29 | d2csua2 |
|
not modelled |
10.4 |
7 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
| 30 | c2d88A_ |
|
not modelled |
10.3 |
24 |
PDB header:signaling protein, protein binding
Chain: A: PDB Molecule:protein mical-3;
PDBTitle: solution structure of the ch domain from human mical-32 protein
|
| 31 | d1ty9a_ |
|
not modelled |
10.0 |
14 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:PNP-oxidase like |
| 32 | d2p8ia1 |
|
not modelled |
9.4 |
14 |
Fold:Ferredoxin-like
Superfamily:DOPA-like
Family:DOPA dioxygenase-like |
| 33 | c2yv2A_ |
|
not modelled |
9.2 |
9 |
PDB header:ligase
Chain: A: PDB Molecule:succinyl-coa synthetase alpha chain;
PDBTitle: crystal structure of succinyl-coa synthetase alpha chain from2 aeropyrum pernix k1
|
| 34 | c3adyA_ |
|
not modelled |
8.9 |
16 |
PDB header:proton transport
Chain: A: PDB Molecule:dotd;
PDBTitle: crystal structure of dotd from legionella
|
| 35 | d1dwka2 |
|
not modelled |
8.8 |
13 |
Fold:Cyanase C-terminal domain
Superfamily:Cyanase C-terminal domain
Family:Cyanase C-terminal domain |
| 36 | c2k1aA_ |
|
not modelled |
8.8 |
29 |
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
|
| 37 | d2f06a2 |
|
not modelled |
8.6 |
15 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:BT0572-like |
| 38 | c2iv1J_ |
|
not modelled |
8.5 |
13 |
PDB header:lyase
Chain: J: PDB Molecule:cyanate hydratase;
PDBTitle: site directed mutagenesis of key residues involved in the2 catalytic mechanism of cyanase
|
| 39 | d1dxxa2 |
|
not modelled |
8.4 |
10 |
Fold:CH domain-like
Superfamily:Calponin-homology domain, CH-domain
Family:Calponin-homology domain, CH-domain |
| 40 | d1sh5a2 |
|
not modelled |
8.3 |
19 |
Fold:CH domain-like
Superfamily:Calponin-homology domain, CH-domain
Family:Calponin-homology domain, CH-domain |
| 41 | c1wylA_ |
|
not modelled |
8.3 |
30 |
PDB header:signaling protein
Chain: A: PDB Molecule:nedd9 interacting protein with calponin homology
PDBTitle: solution structure of the ch domain of human nedd92 interacting protein with calponin homology and lim domains
|
| 42 | d2a2ja1 |
|
not modelled |
8.2 |
15 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:PNP-oxidase like |
| 43 | c2kncB_ |
|
not modelled |
7.9 |
14 |
PDB header:cell adhesion
Chain: B: PDB Molecule:integrin beta-3;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
|
| 44 | c1x3uA_ |
|
not modelled |
7.7 |
26 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulatory protein fixj;
PDBTitle: solution structure of the c-terminal transcriptional2 activator domain of fixj from sinorhizobium melilot
|
| 45 | c2a2jA_ |
|
not modelled |
7.6 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyridoxamine 5'-phosphate oxidase;
PDBTitle: crystal structure of a putative pyridoxine 5'-phosphate oxidase2 (rv2607) from mycobacterium tuberculosis
|
| 46 | d1ku3a_ |
|
not modelled |
7.6 |
12 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
| 47 | c2pebB_ |
|
not modelled |
7.4 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative dioxygenase;
PDBTitle: crystal structure of a putative dioxygenase (npun_f1925) from nostoc2 punctiforme pcc 73102 at 1.46 a resolution
|
| 48 | d3e9va1 |
|
not modelled |
7.4 |
14 |
Fold:BTG domain-like
Superfamily:BTG domain-like
Family:BTG domain-like |
| 49 | c2d87A_ |
|
not modelled |
7.2 |
15 |
PDB header:structural protein, protein binding
Chain: A: PDB Molecule:smoothelin splice isoform l2;
PDBTitle: solution structure of the ch domain from human smoothelin2 splice isoform l2
|
| 50 | d1wb9a4 |
|
not modelled |
7.2 |
13 |
Fold:MutS N-terminal domain-like
Superfamily:DNA repair protein MutS, domain I
Family:DNA repair protein MutS, domain I |
| 51 | c3smtA_ |
|
not modelled |
7.2 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:histone-lysine n-methyltransferase setd3;
PDBTitle: crystal structure of human set domain-containing protein3
|
| 52 | d1bkra_ |
|
not modelled |
7.2 |
15 |
Fold:CH domain-like
Superfamily:Calponin-homology domain, CH-domain
Family:Calponin-homology domain, CH-domain |
| 53 | d1ewqa4 |
|
not modelled |
7.1 |
18 |
Fold:MutS N-terminal domain-like
Superfamily:DNA repair protein MutS, domain I
Family:DNA repair protein MutS, domain I |
| 54 | c3cwbC_ |
|
not modelled |
6.8 |
11 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:cytochrome b;
PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
|
| 55 | d2z15a1 |
|
not modelled |
6.8 |
14 |
Fold:BTG domain-like
Superfamily:BTG domain-like
Family:BTG domain-like |
| 56 | c3hilB_ |
|
not modelled |
6.7 |
38 |
PDB header:transferase
Chain: B: PDB Molecule:ephrin type-a receptor 1;
PDBTitle: sam domain of human ephrin type-a receptor 1 (epha1)
|
| 57 | d3dhxa1 |
|
not modelled |
6.5 |
22 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:NIL domain-like |
| 58 | d1ygya3 |
|
not modelled |
6.5 |
12 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain |
| 59 | d2af7a1 |
|
not modelled |
6.3 |
6 |
Fold:AhpD-like
Superfamily:AhpD-like
Family:CMD-like |
| 60 | c2ogfD_ |
|
not modelled |
6.2 |
12 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:hypothetical protein mj0408;
PDBTitle: crystal structure of protein mj0408 from methanococcus jannaschii,2 pfam duf372
|
| 61 | d1ulra_ |
|
not modelled |
6.2 |
17 |
Fold:Ferredoxin-like
Superfamily:Acylphosphatase/BLUF domain-like
Family:Acylphosphatase-like |
| 62 | c3f6oB_ |
|
not modelled |
5.9 |
18 |
PDB header:transcription regulator
Chain: B: PDB Molecule:probable transcriptional regulator, arsr family
PDBTitle: crystal structure of arsr family transcriptional regulator,2 rha00566
|
| 63 | d2iw0a1 |
|
not modelled |
5.9 |
6 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:NodB-like polysaccharide deacetylase |
| 64 | c2d1uA_ |
|
not modelled |
5.8 |
10 |
PDB header:metal transport
Chain: A: PDB Molecule:iron(iii) dicitrate transport protein feca;
PDBTitle: solution strcuture of the periplasmic signaling domain of2 feca from escherichia coli
|
| 65 | d1ku7a_ |
|
not modelled |
5.6 |
12 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
| 66 | c3mgjA_ |
|
not modelled |
5.6 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein mj1480;
PDBTitle: crystal structure of the saccharop_dh_n domain of mj14802 protein from methanococcus jannaschii. northeast structural3 genomics consortium target mjr83a.
|
| 67 | c3dmyA_ |
|
not modelled |
5.6 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein fdra;
PDBTitle: crystal structure of a predicated acyl-coa synthetase from e.coli
|
| 68 | c2e8nA_ |
|
not modelled |
5.4 |
33 |
PDB header:transferase, signaling protein
Chain: A: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: solution structure of the c-terminal sam-domain of ephaa2:2 ephrin type-a receptor 2 precursor (ec 2.7.10.1)
|
| 69 | d1euca2 |
|
not modelled |
5.2 |
11 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
| 70 | c2yh5A_ |
|
not modelled |
5.1 |
25 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:dapx protein;
PDBTitle: structure of the c-terminal domain of bamc
|
| 71 | c1zzvA_ |
|
not modelled |
5.1 |
10 |
PDB header:membrane protein, metal transport
Chain: A: PDB Molecule:iron(iii) dicitrate transport protein feca;
PDBTitle: solution nmr structure of the periplasmic signaling domain2 of the outer membrane iron transporter feca from3 escherichia coli.
|
| 72 | d1bhda_ |
|
not modelled |
5.0 |
10 |
Fold:CH domain-like
Superfamily:Calponin-homology domain, CH-domain
Family:Calponin-homology domain, CH-domain |