| 1 |
|
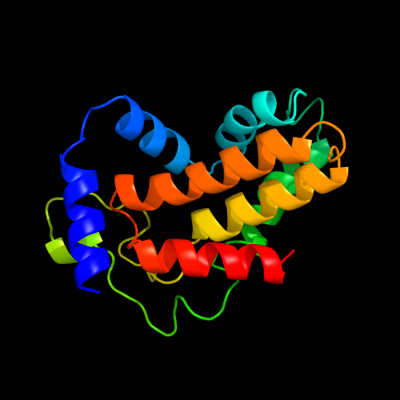
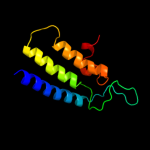
PDB 1d2t chain A
Region: 3 - 165
Aligned: 162
Modelled: 163
Confidence: 99.8%
Identity: 10%
Fold: Acid phosphatase/Vanadium-dependent haloperoxidase
Superfamily: Acid phosphatase/Vanadium-dependent haloperoxidase
Family: Type 2 phosphatidic acid phosphatase, PAP2
Phyre2
| 2 |
|
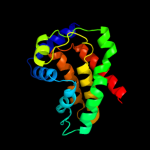
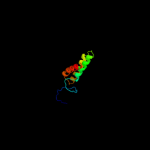
PDB 2akc chain C
Region: 3 - 161
Aligned: 158
Modelled: 159
Confidence: 99.8%
Identity: 18%
PDB header:hydrolase
Chain: C: PDB Molecule:class a nonspecific acid phosphatase phon;
PDBTitle: crystal structure of tungstate complex of the phon protein2 from s. typhimurium
Phyre2
| 3 |
|
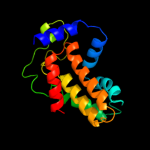
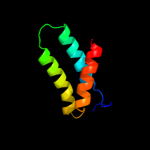
PDB 1qi9 chain A
Region: 79 - 162
Aligned: 84
Modelled: 84
Confidence: 98.3%
Identity: 27%
Fold: Acid phosphatase/Vanadium-dependent haloperoxidase
Superfamily: Acid phosphatase/Vanadium-dependent haloperoxidase
Family: Haloperoxidase (bromoperoxidase)
Phyre2
| 4 |
|
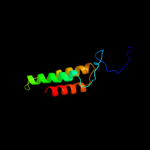
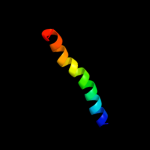
PDB 1vns chain A
Region: 55 - 158
Aligned: 104
Modelled: 104
Confidence: 98.0%
Identity: 26%
Fold: Acid phosphatase/Vanadium-dependent haloperoxidase
Superfamily: Acid phosphatase/Vanadium-dependent haloperoxidase
Family: Chloroperoxidase
Phyre2
| 5 |
|
PDB 1vng chain A
Region: 55 - 158
Aligned: 104
Modelled: 104
Confidence: 97.9%
Identity: 25%
PDB header:haloperoxidase
Chain: A: PDB Molecule:vanadium chloroperoxidase;
PDBTitle: chloroperoxidase from the fungus curvularia inaequalis:2 mutant h404a
Phyre2
| 6 |
|
PDB 1up8 chain A
Region: 79 - 162
Aligned: 84
Modelled: 84
Confidence: 97.5%
Identity: 21%
Fold: Acid phosphatase/Vanadium-dependent haloperoxidase
Superfamily: Acid phosphatase/Vanadium-dependent haloperoxidase
Family: Haloperoxidase (bromoperoxidase)
Phyre2
| 7 |
|
PDB 1qhb chain A
Region: 95 - 162
Aligned: 68
Modelled: 68
Confidence: 97.3%
Identity: 24%
Fold: Acid phosphatase/Vanadium-dependent haloperoxidase
Superfamily: Acid phosphatase/Vanadium-dependent haloperoxidase
Family: Haloperoxidase (bromoperoxidase)
Phyre2
| 8 |
|
PDB 2knc chain B
Region: 150 - 178
Aligned: 29
Modelled: 29
Confidence: 13.7%
Identity: 10%
PDB header:cell adhesion
Chain: B: PDB Molecule:integrin beta-3;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 9 |
|
PDB 2cg4 chain B
Region: 1 - 15
Aligned: 15
Modelled: 15
Confidence: 8.6%
Identity: 13%
PDB header:transcription
Chain: B: PDB Molecule:regulatory protein asnc;
PDBTitle: structure of e.coli asnc
Phyre2
| 10 |
|
PDB 1eh9 chain A domain 2
Region: 97 - 103
Aligned: 7
Modelled: 7
Confidence: 8.1%
Identity: 43%
Fold: Glycosyl hydrolase domain
Superfamily: Glycosyl hydrolase domain
Family: alpha-Amylases, C-terminal beta-sheet domain
Phyre2
| 11 |
|
PDB 2l34 chain A
Region: 150 - 175
Aligned: 26
Modelled: 25
Confidence: 7.7%
Identity: 15%
PDB header:protein binding
Chain: A: PDB Molecule:tyro protein tyrosine kinase-binding protein;
PDBTitle: structure of the dap12 transmembrane homodimer
Phyre2
| 12 |
|
PDB 2l34 chain B
Region: 150 - 175
Aligned: 26
Modelled: 25
Confidence: 7.7%
Identity: 15%
PDB header:protein binding
Chain: B: PDB Molecule:tyro protein tyrosine kinase-binding protein;
PDBTitle: structure of the dap12 transmembrane homodimer
Phyre2
| 13 |
|
PDB 2cg4 chain A domain 1
Region: 1 - 20
Aligned: 20
Modelled: 20
Confidence: 6.5%
Identity: 15%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: Lrp/AsnC-like transcriptional regulator N-terminal domain
Phyre2
| 14 |
|
PDB 1dsv chain A
Region: 143 - 149
Aligned: 7
Modelled: 7
Confidence: 5.6%
Identity: 43%
Fold: Retrovirus zinc finger-like domains
Superfamily: Retrovirus zinc finger-like domains
Family: Retrovirus zinc finger-like domains
Phyre2
| 15 |
|
PDB 2r4q chain A domain 1
Region: 100 - 110
Aligned: 11
Modelled: 10
Confidence: 5.5%
Identity: 18%
Fold: Phosphotyrosine protein phosphatases I-like
Superfamily: PTS system IIB component-like
Family: PTS system, Fructose specific IIB subunit-like
Phyre2