| 1 |
|
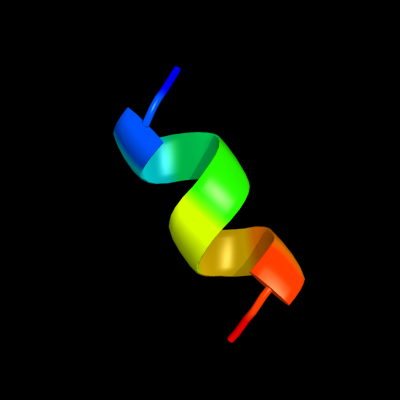
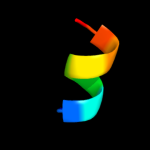
PDB 2y94 chain C
Region: 42 - 51
Aligned: 10
Modelled: 10
Confidence: 13.6%
Identity: 60%
PDB header:transferase
Chain: C: PDB Molecule:5'-amp-activated protein kinase catalytic subunit alpha-1;
PDBTitle: structure of an active form of mammalian ampk
Phyre2
| 2 |
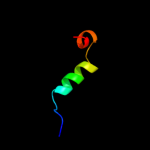
|
PDB 1ejx chain B
Region: 5 - 19
Aligned: 15
Modelled: 15
Confidence: 12.3%
Identity: 27%
Fold: beta-clip
Superfamily: Urease, beta-subunit
Family: Urease, beta-subunit
Phyre2
| 3 |
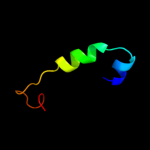
|
PDB 4ubp chain B
Region: 5 - 19
Aligned: 15
Modelled: 15
Confidence: 12.0%
Identity: 40%
Fold: beta-clip
Superfamily: Urease, beta-subunit
Family: Urease, beta-subunit
Phyre2
| 4 |
|
PDB 3qb2 chain A
Region: 38 - 67
Aligned: 30
Modelled: 30
Confidence: 11.4%
Identity: 23%
PDB header:hydrolase inhibitor
Chain: A: PDB Molecule:immunity factor for spn;
PDBTitle: the crystal structure of immunity factor for spn (ifs)
Phyre2
| 5 |
|
PDB 1e9y chain A domain 1
Region: 5 - 19
Aligned: 15
Modelled: 15
Confidence: 10.3%
Identity: 33%
Fold: beta-clip
Superfamily: Urease, beta-subunit
Family: Urease, beta-subunit
Phyre2
| 6 |
|
PDB 3izx chain E
Region: 59 - 83
Aligned: 25
Modelled: 25
Confidence: 9.6%
Identity: 28%
PDB header:virus
Chain: E: PDB Molecule:viral structural protein 5;
PDBTitle: 3.1 angstrom cryoem structure of cytoplasmic polyhedrosis virus
Phyre2
| 7 |
|
PDB 2i5n chain H domain 2
Region: 24 - 32
Aligned: 9
Modelled: 9
Confidence: 9.3%
Identity: 44%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction centre subunit H, transmembrane region
Family: Photosystem II reaction centre subunit H, transmembrane region
Phyre2
| 8 |
|
PDB 3iz3 chain D
Region: 59 - 83
Aligned: 25
Modelled: 25
Confidence: 9.1%
Identity: 28%
PDB header:virus
Chain: D: PDB Molecule:viral structural protein 5;
PDBTitle: cryoem structure of cytoplasmic polyhedrosis virus
Phyre2
| 9 |
|
PDB 3pnt chain D
Region: 38 - 67
Aligned: 30
Modelled: 30
Confidence: 8.4%
Identity: 23%
PDB header:hydrolase/hydrolase inhibitor
Chain: D: PDB Molecule:immunity factor for spn;
PDBTitle: crystal structure of the streptococcus pyogenes nad+ glycohydrolase2 spn in complex with ifs, the immunity factor for spn
Phyre2
| 10 |
|
PDB 2xb0 chain X
Region: 18 - 34
Aligned: 17
Modelled: 17
Confidence: 8.1%
Identity: 12%
PDB header:hydrolase
Chain: X: PDB Molecule:chromo domain-containing protein 1;
PDBTitle: dna-binding domain from saccharomyces cerevisiae chromatin-2 remodelling protein chd1
Phyre2
| 11 |
|
PDB 2e9x chain F
Region: 40 - 66
Aligned: 27
Modelled: 27
Confidence: 6.1%
Identity: 22%
PDB header:replication
Chain: F: PDB Molecule:dna replication complex gins protein psf2;
PDBTitle: the crystal structure of human gins core complex
Phyre2
| 12 |
|
PDB 3qga chain D
Region: 5 - 19
Aligned: 15
Modelled: 15
Confidence: 5.9%
Identity: 40%
PDB header:hydrolase
Chain: D: PDB Molecule:fusion of urease beta and gamma subunits;
PDBTitle: 3.0 a model of iron containing urease urea2b2 from helicobacter2 mustelae
Phyre2
| 13 |
|
PDB 1l3a chain C
Region: 8 - 25
Aligned: 18
Modelled: 18
Confidence: 5.7%
Identity: 17%
Fold: ssDNA-binding transcriptional regulator domain
Superfamily: ssDNA-binding transcriptional regulator domain
Family: Plant transcriptional regulator PBF-2
Phyre2