| 1 |
|
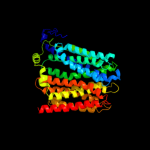
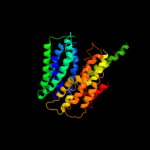
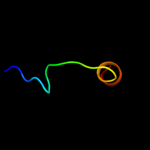
PDB 1pw4 chain A
Region: 6 - 463
Aligned: 420
Modelled: 435
Confidence: 100.0%
Identity: 14%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 2 |
|
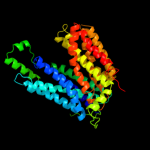
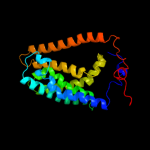
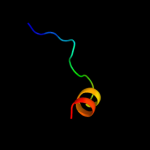
PDB 3o7p chain A
Region: 36 - 461
Aligned: 390
Modelled: 397
Confidence: 100.0%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
Phyre2
| 3 |
|
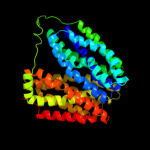
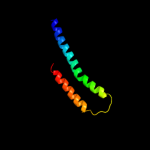
PDB 1pv7 chain A
Region: 31 - 462
Aligned: 403
Modelled: 404
Confidence: 100.0%
Identity: 11%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 4 |
|
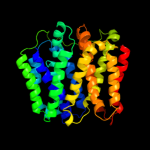
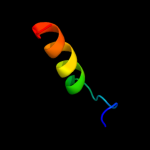
PDB 2gfp chain A
Region: 38 - 448
Aligned: 366
Modelled: 372
Confidence: 100.0%
Identity: 8%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 5 |
|
PDB 2xut chain C
Region: 39 - 452
Aligned: 410
Modelled: 414
Confidence: 100.0%
Identity: 10%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 6 |
|
PDB 3b9y chain A
Region: 23 - 262
Aligned: 229
Modelled: 240
Confidence: 70.0%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
Phyre2
| 7 |
|
PDB 3hd6 chain A
Region: 12 - 246
Aligned: 219
Modelled: 227
Confidence: 52.1%
Identity: 13%
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
Phyre2
| 8 |
|
PDB 2e8p chain A
Region: 2 - 28
Aligned: 27
Modelled: 27
Confidence: 18.5%
Identity: 33%
PDB header:signaling protein
Chain: A: PDB Molecule:elf3 protein;
PDBTitle: solution structure of the n-terminal sam-domain of e74-like2 factor 3
Phyre2
| 9 |
|
PDB 2rdd chain B
Region: 440 - 463
Aligned: 24
Modelled: 24
Confidence: 10.5%
Identity: 13%
PDB header:membrane protein/transport protein
Chain: B: PDB Molecule:upf0092 membrane protein yajc;
PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
Phyre2
| 10 |
|
PDB 2l92 chain A
Region: 19 - 35
Aligned: 17
Modelled: 17
Confidence: 9.4%
Identity: 29%
PDB header:dna binding protein
Chain: A: PDB Molecule:histone family protein nucleoid-structuring protein h-ns;
PDBTitle: solution structure of the c-terminal domain of h-ns like protein bv3f
Phyre2
| 11 |
|
PDB 2g9p chain A
Region: 91 - 104
Aligned: 14
Modelled: 14
Confidence: 7.9%
Identity: 14%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:antimicrobial peptide latarcin 2a;
PDBTitle: nmr structure of a novel antimicrobial peptide, latarcin 2a,2 from spider (lachesana tarabaevi) venom
Phyre2
| 12 |
|
PDB 2bbj chain B
Region: 394 - 460
Aligned: 67
Modelled: 67
Confidence: 7.0%
Identity: 12%
PDB header:metal transport/membrane protein
Chain: B: PDB Molecule:divalent cation transport-related protein;
PDBTitle: crystal structure of the cora mg2+ transporter
Phyre2
| 13 |
|
PDB 3jyv chain T
Region: 20 - 40
Aligned: 21
Modelled: 21
Confidence: 5.9%
Identity: 19%
PDB header:ribosome
Chain: T: PDB Molecule:s19e protein;
PDBTitle: structure of the 40s rrna and proteins and p/e trna for eukaryotic2 ribosome based on cryo-em map of thermomyces lanuginosus ribosome at3 8.9a resolution
Phyre2
| 14 |
|
PDB 3lge chain G
Region: 13 - 31
Aligned: 19
Modelled: 18
Confidence: 5.8%
Identity: 16%
PDB header:lyase/protein binding
Chain: G: PDB Molecule:sorting nexin-9;
PDBTitle: crystal structure of rabbit muscle aldolase-snx9 lc4 complex
Phyre2
| 15 |
|
PDB 3lge chain F
Region: 13 - 31
Aligned: 19
Modelled: 18
Confidence: 5.8%
Identity: 16%
PDB header:lyase/protein binding
Chain: F: PDB Molecule:sorting nexin-9;
PDBTitle: crystal structure of rabbit muscle aldolase-snx9 lc4 complex
Phyre2
| 16 |
|
PDB 2inp chain D
Region: 92 - 103
Aligned: 12
Modelled: 12
Confidence: 5.5%
Identity: 25%
PDB header:oxidoreductase
Chain: D: PDB Molecule:phenol hydroxylase component phl;
PDBTitle: structure of the phenol hydroxylase-regulatory protein2 complex
Phyre2
| 17 |
|
PDB 2imh chain A domain 1
Region: 7 - 23
Aligned: 17
Modelled: 17
Confidence: 5.4%
Identity: 29%
Fold: Ntn hydrolase-like
Superfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)
Family: SPO2555-like
Phyre2
| 18 |
|
PDB 3hft chain A
Region: 13 - 29
Aligned: 17
Modelled: 17
Confidence: 5.2%
Identity: 35%
PDB header:hydrolase
Chain: A: PDB Molecule:wbms, polysaccharide deacetylase involved in o-antigen
PDBTitle: crystal structure of a putative polysaccharide deacetylase involved in2 o-antigen biosynthesis (wbms, bb0128) from bordetella bronchiseptica3 at 1.90 a resolution
Phyre2