| 1 |
|
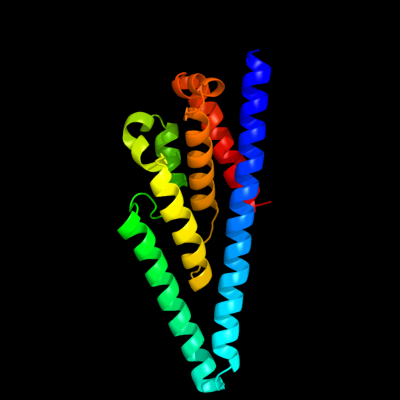
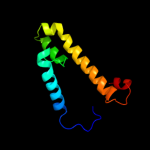
PDB 3org chain B
Region: 174 - 341
Aligned: 168
Modelled: 168
Confidence: 52.3%
Identity: 11%
PDB header:transport protein
Chain: B: PDB Molecule:cmclc;
PDBTitle: crystal structure of a eukaryotic clc transporter
Phyre2
| 2 |
|
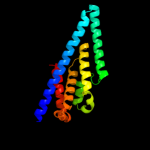
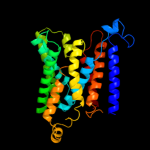
PDB 1y5i chain C domain 1
Region: 217 - 342
Aligned: 126
Modelled: 126
Confidence: 27.9%
Identity: 10%
Fold: Heme-binding four-helical bundle
Superfamily: Respiratory nitrate reductase 1 gamma chain
Family: Respiratory nitrate reductase 1 gamma chain
Phyre2
| 3 |
|
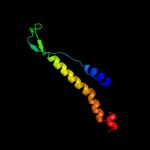
PDB 1w8x chain P
Region: 1 - 21
Aligned: 21
Modelled: 21
Confidence: 24.6%
Identity: 24%
PDB header:virus
Chain: P: PDB Molecule:protein p16;
PDBTitle: structural analysis of prd1
Phyre2
| 4 |
|
PDB 2w8a chain C
Region: 125 - 266
Aligned: 118
Modelled: 124
Confidence: 13.1%
Identity: 9%
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
Phyre2
| 5 |
|
PDB 1ymg chain A
Region: 167 - 259
Aligned: 89
Modelled: 93
Confidence: 11.9%
Identity: 18%
PDB header:membrane protein
Chain: A: PDB Molecule:lens fiber major intrinsic protein;
PDBTitle: the channel architecture of aquaporin o at 2.2 angstrom resolution
Phyre2
| 6 |
|
PDB 1ymg chain A domain 1
Region: 167 - 259
Aligned: 89
Modelled: 93
Confidence: 11.9%
Identity: 18%
Fold: Aquaporin-like
Superfamily: Aquaporin-like
Family: Aquaporin-like
Phyre2
| 7 |
|
PDB 3d9s chain B
Region: 167 - 263
Aligned: 94
Modelled: 97
Confidence: 10.4%
Identity: 13%
PDB header:membrane protein
Chain: B: PDB Molecule:aquaporin-5;
PDBTitle: human aquaporin 5 (aqp5) - high resolution x-ray structure
Phyre2
| 8 |
|
PDB 1j4n chain A
Region: 167 - 256
Aligned: 87
Modelled: 90
Confidence: 9.2%
Identity: 11%
Fold: Aquaporin-like
Superfamily: Aquaporin-like
Family: Aquaporin-like
Phyre2
| 9 |
|
PDB 1l7v chain A
Region: 49 - 343
Aligned: 288
Modelled: 284
Confidence: 9.2%
Identity: 11%
Fold: ABC transporter involved in vitamin B12 uptake, BtuC
Superfamily: ABC transporter involved in vitamin B12 uptake, BtuC
Family: ABC transporter involved in vitamin B12 uptake, BtuC
Phyre2
| 10 |
|
PDB 3qnq chain D
Region: 222 - 258
Aligned: 37
Modelled: 37
Confidence: 8.7%
Identity: 5%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 11 |
|
PDB 1fft chain B domain 2
Region: 198 - 285
Aligned: 78
Modelled: 88
Confidence: 7.7%
Identity: 13%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 12 |
|
PDB 2oar chain A
Region: 175 - 260
Aligned: 86
Modelled: 86
Confidence: 7.5%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:large-conductance mechanosensitive channel;
PDBTitle: mechanosensitive channel of large conductance (mscl)
Phyre2