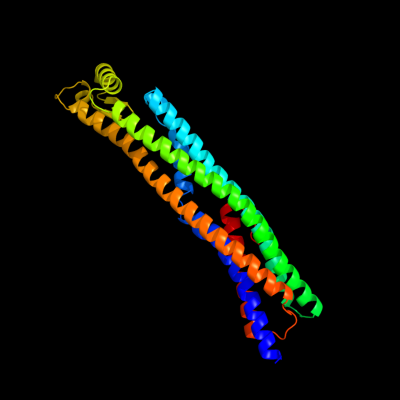
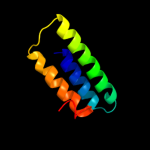
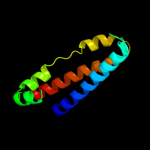
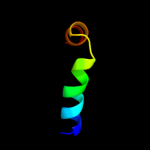
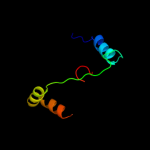
1 c1qoyA_
100.0
99
PDB header: toxinChain: A: PDB Molecule: hemolysin e;PDBTitle: e.coli hemolysin e (hlye, clya, shea)
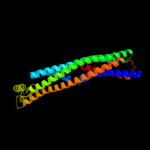
2 c3brvB_
20.7
6
PDB header: transferase/transcriptionChain: B: PDB Molecule: nf-kappa-b essential modulator;PDBTitle: nemo/ikkb association domain structure
3 d1pcaa1
17.1
14
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Pancreatic carboxypeptidase, activation domain4 d2coba1
15.3
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Psq domain5 d1r11a3
14.0
29
Fold: MutS N-terminal domain-likeSuperfamily: tRNA-intron endonuclease N-terminal domain-likeFamily: tRNA-intron endonuclease N-terminal domain-like6 c3pjaK_
13.8
15
PDB header: hydrolaseChain: K: PDB Molecule: translin-associated protein x;PDBTitle: crystal structure of human c3po complex
7 c2q1kA_
12.6
14
PDB header: chaperoneChain: A: PDB Molecule: asce;PDBTitle: cyrstal structure of asce from aeromonas hydrophilla
8 d1f6fa_
12.4
10
Fold: 4-helical cytokinesSuperfamily: 4-helical cytokinesFamily: Long-chain cytokines9 d1pfva1
12.0
16
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesSuperfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesFamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases10 c4a9zD_
11.2
13
PDB header: transcriptionChain: D: PDB Molecule: tumor protein 63;PDBTitle: crystal structure of human p63 tetramerization domain
11 d1rqga1
10.5
16
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesSuperfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesFamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases12 d1gg2g_
10.3
23
Fold: Non-globular all-alpha subunits of globular proteinsSuperfamily: Transducin (heterotrimeric G protein), gamma chainFamily: Transducin (heterotrimeric G protein), gamma chain13 c3f1iH_
10.1
13
PDB header: protein bindingChain: H: PDB Molecule: hepatocyte growth factor-regulated tyrosine kinasePDBTitle: human escrt-0 core complex
14 d1h3na1
10.1
14
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesSuperfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesFamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases15 d1rw5a1
9.9
7
Fold: 4-helical cytokinesSuperfamily: 4-helical cytokinesFamily: Long-chain cytokines16 c3zy1A_
9.9
13
PDB header: transcriptionChain: A: PDB Molecule: tumor protein 63;PDBTitle: crystal structure of the human p63 tetramerization domain
17 c3kwlA_
9.4
18
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a hypothetical protein from helicobacter pylori
18 c1ciiA_
9.4
13
PDB header: transmembrane proteinChain: A: PDB Molecule: colicin ia;PDBTitle: colicin ia
19 c2ql2A_
8.9
23
PDB header: transcription/dnaChain: A: PDB Molecule: transcription factor e2-alpha;PDBTitle: crystal structure of the basic-helix-loop-helix domains of2 the heterodimer e47/neurod1 bound to dna
20 d2ezwa1
8.9
27
Fold: Dimerization-anchoring domain of cAMP-dependent PK regulatory subunitSuperfamily: Dimerization-anchoring domain of cAMP-dependent PK regulatory subunitFamily: Dimerization-anchoring domain of cAMP-dependent PK regulatory subunit21 d1b0na1
not modelled
8.5
12
Fold: Dimerisation interlockSuperfamily: SinR repressor dimerisation domain-likeFamily: SinR repressor dimerisation domain-like22 d1omwg_
not modelled
8.4
17
Fold: Non-globular all-alpha subunits of globular proteinsSuperfamily: Transducin (heterotrimeric G protein), gamma chainFamily: Transducin (heterotrimeric G protein), gamma chain23 c2k29A_
not modelled
8.1
24
PDB header: transcriptionChain: A: PDB Molecule: antitoxin relb;PDBTitle: structure of the dbd domain of e. coli antitoxin relb
24 d1ggpa_
not modelled
7.7
16
Fold: Ribosome inactivating proteins (RIP)Superfamily: Ribosome inactivating proteins (RIP)Family: Plant cytotoxins25 d1gotg_
not modelled
7.2
9
Fold: Non-globular all-alpha subunits of globular proteinsSuperfamily: Transducin (heterotrimeric G protein), gamma chainFamily: Transducin (heterotrimeric G protein), gamma chain26 d1xppa_
not modelled
7.2
8
Fold: DCoH-likeSuperfamily: RBP11-like subunits of RNA polymeraseFamily: RBP11/RpoL27 d1tbge_
not modelled
6.3
9
Fold: Non-globular all-alpha subunits of globular proteinsSuperfamily: Transducin (heterotrimeric G protein), gamma chainFamily: Transducin (heterotrimeric G protein), gamma chain28 c1gaxB_
not modelled
6.2
15
PDB header: ligase/rnaChain: B: PDB Molecule: valyl-trna synthetase;PDBTitle: crystal structure of thermus thermophilus valyl-trna2 synthetase complexed with trna(val) and valyl-adenylate3 analogue
29 c2zwmA_
not modelled
6.1
14
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulatory protein yycf;PDBTitle: crystal structure of yycf receiver domain from bacillus2 subtilis
30 c2rlwA_
not modelled
5.9
16
PDB header: toxinChain: A: PDB Molecule: plnf;PDBTitle: three-dimensional structure of the two peptides that2 constitute the two-peptide bacteriocin plantaracin ef
31 c3qqwC_
not modelled
5.9
19
PDB header: lyaseChain: C: PDB Molecule: putative citrate lyase;PDBTitle: crystal structure of a hypothetical lyase (reut_b4148) from ralstonia2 eutropha jmp134 at 2.44 a resolution
32 c3hisA_
not modelled
5.9
11
PDB header: hydrolaseChain: A: PDB Molecule: vacuolar saporin;PDBTitle: crystal structure of saporin-l1 from saponaria officinalis
33 c3df8A_
not modelled
5.8
12
PDB header: transcriptionChain: A: PDB Molecule: possible hxlr family transcriptional factor;PDBTitle: the crystal structure of a possible hxlr family transcriptional factor2 from thermoplasma volcanium gss1
34 c2xzmV_
not modelled
5.7
20
PDB header: ribosomeChain: V: PDB Molecule: rps17e;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
35 d1vcsa1
not modelled
5.6
14
Fold: STAT-likeSuperfamily: t-snare proteinsFamily: t-snare proteins36 c1jsdB_
not modelled
5.5
23
PDB header: viral proteinChain: B: PDB Molecule: haemagglutinin (ha2 chain);PDBTitle: crystal structure of swine h9 haemagglutinin
37 c1lt1G_
not modelled
5.4
11
PDB header: de novo proteinChain: G: PDB Molecule: l13g-df1;PDBTitle: sliding helix induced change of coordination geometry in a2 model di-mn(ii) protein
38 c3i5pA_
not modelled
5.4
18
PDB header: protein transportChain: A: PDB Molecule: nucleoporin nup170;PDBTitle: nup170(aa979-1502), s.cerevisiae
39 c2uwjE_
not modelled
5.3
11
PDB header: chaperoneChain: E: PDB Molecule: type iii export protein psce;PDBTitle: structure of the heterotrimeric complex which regulates2 type iii secretion needle formation
40 d1d9ca_
not modelled
5.3
16
Fold: 4-helical cytokinesSuperfamily: 4-helical cytokinesFamily: Interferons/interleukin-10 (IL-10)41 d1twfk_
not modelled
5.1
16
Fold: DCoH-likeSuperfamily: RBP11-like subunits of RNA polymeraseFamily: RBP11/RpoL