| 1 |
|
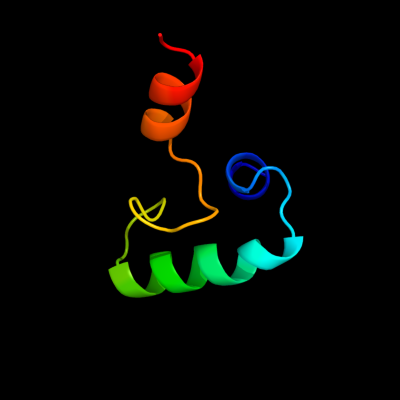
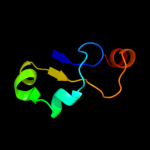
PDB 3cz6 chain A
Region: 142 - 208
Aligned: 47
Modelled: 47
Confidence: 14.6%
Identity: 28%
PDB header:protein binding
Chain: A: PDB Molecule:dna-binding protein rap1;
PDBTitle: crystal structure of the rap1 c-terminus
Phyre2
| 2 |
|
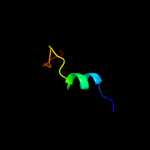
PDB 2qqh chain A
Region: 296 - 329
Aligned: 28
Modelled: 34
Confidence: 12.6%
Identity: 25%
PDB header:immune system, membrane protein
Chain: A: PDB Molecule:complement component c8 alpha chain;
PDBTitle: structure of c8a-macpf reveals mechanism of membrane attack2 in complement immune defense
Phyre2
| 3 |
|
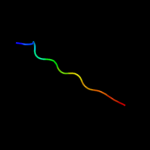
PDB 1bg6 chain A domain 1
Region: 192 - 213
Aligned: 22
Modelled: 22
Confidence: 12.1%
Identity: 23%
Fold: 6-phosphogluconate dehydrogenase C-terminal domain-like
Superfamily: 6-phosphogluconate dehydrogenase C-terminal domain-like
Family: N-(1-D-carboxylethyl)-L-norvaline dehydrogenase
Phyre2
| 4 |
|
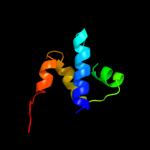
PDB 1es9 chain A
Region: 286 - 331
Aligned: 44
Modelled: 46
Confidence: 9.8%
Identity: 20%
Fold: Flavodoxin-like
Superfamily: SGNH hydrolase
Family: Acetylhydrolase
Phyre2
| 5 |
|
PDB 3h0g chain E
Region: 272 - 284
Aligned: 13
Modelled: 13
Confidence: 7.6%
Identity: 38%
PDB header:transcription
Chain: E: PDB Molecule:dna-directed rna polymerases i, ii, and iii
PDBTitle: rna polymerase ii from schizosaccharomyces pombe
Phyre2
| 6 |
|
PDB 1dzf chain A domain 2
Region: 273 - 283
Aligned: 11
Modelled: 11
Confidence: 7.4%
Identity: 45%
Fold: RPB5-like RNA polymerase subunit
Superfamily: RPB5-like RNA polymerase subunit
Family: RPB5
Phyre2
| 7 |
|
PDB 1n8i chain A
Region: 301 - 325
Aligned: 25
Modelled: 25
Confidence: 7.3%
Identity: 36%
Fold: TIM beta/alpha-barrel
Superfamily: Malate synthase G
Family: Malate synthase G
Phyre2
| 8 |
|
PDB 2f22 chain A domain 1
Region: 189 - 200
Aligned: 12
Modelled: 12
Confidence: 6.6%
Identity: 50%
Fold: DinB/YfiT-like putative metalloenzymes
Superfamily: DinB/YfiT-like putative metalloenzymes
Family: DinB-like
Phyre2
| 9 |
|
PDB 1hmj chain A
Region: 273 - 283
Aligned: 11
Modelled: 11
Confidence: 6.3%
Identity: 55%
Fold: RPB5-like RNA polymerase subunit
Superfamily: RPB5-like RNA polymerase subunit
Family: RPB5
Phyre2
| 10 |
|
PDB 2lbf chain A
Region: 2 - 64
Aligned: 63
Modelled: 63
Confidence: 5.6%
Identity: 29%
PDB header:ribosomal protein
Chain: A: PDB Molecule:60s acidic ribosomal protein p1;
PDBTitle: solution structure of the dimerization domain of human ribosomal2 protein p1/p2 heterodimer
Phyre2
| 11 |
|
PDB 2axt chain A domain 1
Region: 333 - 385
Aligned: 51
Modelled: 53
Confidence: 5.4%
Identity: 24%
Fold: Bacterial photosystem II reaction centre, L and M subunits
Superfamily: Bacterial photosystem II reaction centre, L and M subunits
Family: Bacterial photosystem II reaction centre, L and M subunits
Phyre2