| 1 |
|
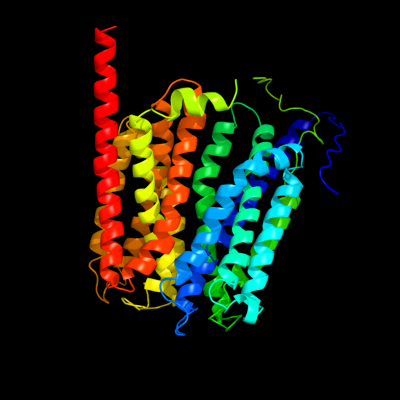
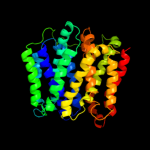
PDB 1pw4 chain A
Region: 5 - 448
Aligned: 431
Modelled: 431
Confidence: 100.0%
Identity: 97%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 2 |
|
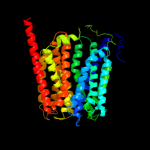
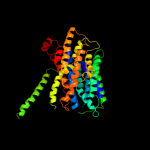
PDB 1pv7 chain A
Region: 19 - 450
Aligned: 414
Modelled: 432
Confidence: 100.0%
Identity: 12%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 3 |
|
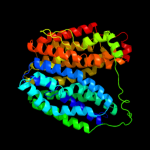
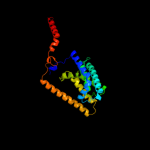
PDB 3o7p chain A
Region: 25 - 438
Aligned: 401
Modelled: 414
Confidence: 100.0%
Identity: 8%
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
Phyre2
| 4 |
|
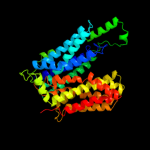
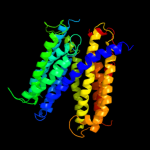
PDB 2gfp chain A
Region: 30 - 428
Aligned: 369
Modelled: 377
Confidence: 100.0%
Identity: 15%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 5 |
|
PDB 2xut chain C
Region: 28 - 434
Aligned: 402
Modelled: 407
Confidence: 100.0%
Identity: 12%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 6 |
|
PDB 3b9y chain A
Region: 16 - 253
Aligned: 228
Modelled: 238
Confidence: 92.5%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
Phyre2
| 7 |
|
PDB 3mku chain A
Region: 15 - 449
Aligned: 433
Modelled: 433
Confidence: 44.4%
Identity: 9%
PDB header:transport protein
Chain: A: PDB Molecule:multi antimicrobial extrusion protein (na(+)/drug
PDBTitle: structure of a cation-bound multidrug and toxin compound extrusion2 (mate) transporter
Phyre2
| 8 |
|
PDB 3qnq chain D
Region: 401 - 451
Aligned: 51
Modelled: 51
Confidence: 40.8%
Identity: 18%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 9 |
|
PDB 3hd6 chain A
Region: 14 - 238
Aligned: 219
Modelled: 225
Confidence: 39.7%
Identity: 10%
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
Phyre2
| 10 |
|
PDB 2rdd chain B
Region: 420 - 448
Aligned: 29
Modelled: 29
Confidence: 26.1%
Identity: 3%
PDB header:membrane protein/transport protein
Chain: B: PDB Molecule:upf0092 membrane protein yajc;
PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
Phyre2
| 11 |
|
PDB 2g9p chain A
Region: 82 - 95
Aligned: 14
Modelled: 14
Confidence: 9.1%
Identity: 7%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:antimicrobial peptide latarcin 2a;
PDBTitle: nmr structure of a novel antimicrobial peptide, latarcin 2a,2 from spider (lachesana tarabaevi) venom
Phyre2
| 12 |
|
PDB 3prr chain T
Region: 414 - 444
Aligned: 31
Modelled: 31
Confidence: 5.8%
Identity: 16%
PDB header:photosynthesis
Chain: T: PDB Molecule:photosystem ii reaction center protein t;
PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 2 of 2). this file contains second monomer of psii3 dimer
Phyre2
| 13 |
|
PDB 3prq chain T
Region: 414 - 444
Aligned: 31
Modelled: 31
Confidence: 5.8%
Identity: 16%
PDB header:photosynthesis
Chain: T: PDB Molecule:photosystem ii reaction center protein t;
PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 1 of 2). this file contains first monomer of psii3 dimer
Phyre2
| 14 |
|
PDB 3bz1 chain T
Region: 414 - 444
Aligned: 31
Modelled: 31
Confidence: 5.8%
Identity: 16%
PDB header:electron transport
Chain: T: PDB Molecule:photosystem ii reaction center protein t;
PDBTitle: crystal structure of cyanobacterial photosystem ii (part 12 of 2). this file contains first monomer of psii dimer
Phyre2
| 15 |
|
PDB 3bz2 chain T
Region: 414 - 444
Aligned: 31
Modelled: 31
Confidence: 5.8%
Identity: 16%
PDB header:electron transport
Chain: T: PDB Molecule:photosystem ii reaction center protein t;
PDBTitle: crystal structure of cyanobacterial photosystem ii (part 22 of 2). this file contains second monomer of psii dimer
Phyre2
| 16 |
|
PDB 3dtu chain B domain 2
Region: 408 - 451
Aligned: 44
Modelled: 44
Confidence: 5.7%
Identity: 7%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 17 |
|
PDB 2knc chain A
Region: 413 - 452
Aligned: 40
Modelled: 40
Confidence: 5.4%
Identity: 23%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 18 |
|
PDB 1k4u chain P
Region: 6 - 19
Aligned: 14
Modelled: 14
Confidence: 5.2%
Identity: 36%
PDB header:hormone/growth factor
Chain: P: PDB Molecule:phagocyte nadph oxidase subunit p47phox;
PDBTitle: solution structure of the c-terminal sh3 domain of p67phox2 complexed with the c-terminal tail region of p47phox
Phyre2
| 19 |
|
PDB 2jyn chain A
Region: 5 - 32
Aligned: 28
Modelled: 28
Confidence: 5.2%
Identity: 18%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0368 protein ypl225w;
PDBTitle: a novel solution nmr structure of protein yst0336 from2 saccharomyces cerevisiae. northeast structural genomics3 consortium target yt51/ontario centre for structural4 proteomics target yst0336
Phyre2