| 1 |
|
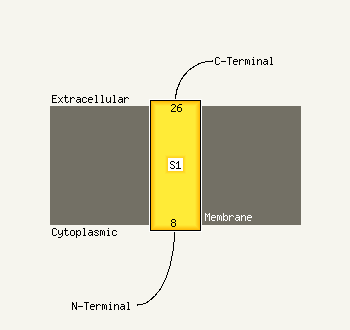
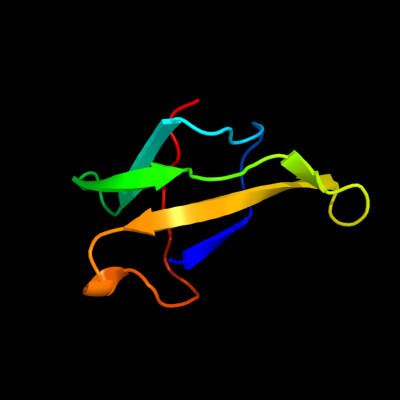
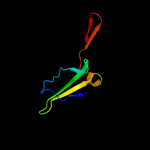
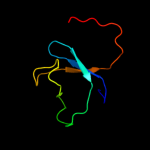
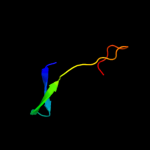
PDB 1v5v chain A domain 1
Region: 41 - 101
Aligned: 61
Modelled: 61
Confidence: 64.8%
Identity: 16%
Fold: Elongation factor/aminomethyltransferase common domain
Superfamily: Aminomethyltransferase beta-barrel domain
Family: Aminomethyltransferase beta-barrel domain
Phyre2
| 2 |
|
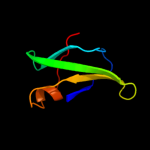
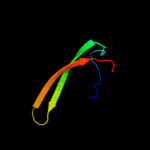
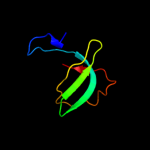
PDB 2j5u chain B
Region: 61 - 93
Aligned: 33
Modelled: 33
Confidence: 63.6%
Identity: 9%
PDB header:cell shape regulation
Chain: B: PDB Molecule:mrec protein;
PDBTitle: mrec lysteria monocytogenes
Phyre2
| 3 |
|
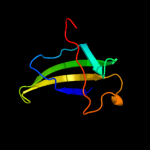
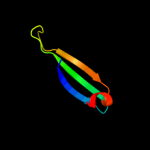
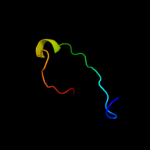
PDB 1v5v chain A
Region: 38 - 105
Aligned: 66
Modelled: 68
Confidence: 40.2%
Identity: 15%
PDB header:transferase
Chain: A: PDB Molecule:aminomethyltransferase;
PDBTitle: crystal structure of a component of glycine cleavage system: t-protein2 from pyrococcus horikoshii ot3 at 1.5 a resolution
Phyre2
| 4 |
|
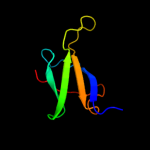
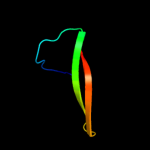
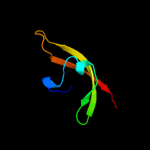
PDB 2qf4 chain A
Region: 41 - 108
Aligned: 63
Modelled: 68
Confidence: 32.2%
Identity: 14%
PDB header:structural protein
Chain: A: PDB Molecule:cell shape determining protein mrec;
PDBTitle: high resolution structure of the major periplasmic domain from the2 cell shape-determining filament mrec (orthorhombic form)
Phyre2
| 5 |
|
PDB 1pj5 chain A domain 1
Region: 42 - 101
Aligned: 59
Modelled: 60
Confidence: 15.2%
Identity: 17%
Fold: Elongation factor/aminomethyltransferase common domain
Superfamily: Aminomethyltransferase beta-barrel domain
Family: Aminomethyltransferase beta-barrel domain
Phyre2
| 6 |
|
PDB 1wos chain A domain 1
Region: 41 - 105
Aligned: 63
Modelled: 65
Confidence: 13.9%
Identity: 10%
Fold: Elongation factor/aminomethyltransferase common domain
Superfamily: Aminomethyltransferase beta-barrel domain
Family: Aminomethyltransferase beta-barrel domain
Phyre2
| 7 |
|
PDB 1yx2 chain B
Region: 36 - 101
Aligned: 66
Modelled: 66
Confidence: 12.9%
Identity: 17%
PDB header:transferase
Chain: B: PDB Molecule:aminomethyltransferase;
PDBTitle: crystal structure of the probable aminomethyltransferase2 from bacillus subtilis
Phyre2
| 8 |
|
PDB 1cz5 chain A
Region: 51 - 116
Aligned: 65
Modelled: 66
Confidence: 11.0%
Identity: 11%
PDB header:hydrolase
Chain: A: PDB Molecule:vcp-like atpase;
PDBTitle: nmr structure of vat-n: the n-terminal domain of vat (vcp-2 like atpase of thermoplasma)
Phyre2
| 9 |
|
PDB 2k6z chain A
Region: 25 - 61
Aligned: 37
Modelled: 37
Confidence: 10.5%
Identity: 16%
PDB header:metal transport
Chain: A: PDB Molecule:putative uncharacterized protein ttha1943;
PDBTitle: solution structures of copper loaded form pcua (trans2 conformation of the peptide bond involving the nitrogen of3 p14)
Phyre2
| 10 |
|
PDB 1h9r chain A domain 2
Region: 66 - 108
Aligned: 36
Modelled: 43
Confidence: 9.2%
Identity: 8%
Fold: OB-fold
Superfamily: MOP-like
Family: BiMOP, duplicated molybdate-binding domain
Phyre2
| 11 |
|
PDB 1x9l chain A
Region: 25 - 59
Aligned: 35
Modelled: 35
Confidence: 8.1%
Identity: 9%
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily: DR1885-like metal-binding protein
Family: DR1885-like metal-binding protein
Phyre2
| 12 |
|
PDB 3htr chain B
Region: 54 - 81
Aligned: 28
Modelled: 28
Confidence: 7.9%
Identity: 11%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized prc-barrel domain protein;
PDBTitle: crystal structure of prc-barrel domain protein from2 rhodopseudomonas palustris
Phyre2
| 13 |
|
PDB 1t5e chain B
Region: 35 - 61
Aligned: 27
Modelled: 27
Confidence: 7.8%
Identity: 22%
PDB header:transport protein
Chain: B: PDB Molecule:multidrug resistance protein mexa;
PDBTitle: the structure of mexa
Phyre2
| 14 |
|
PDB 1nnx chain A
Region: 48 - 102
Aligned: 55
Modelled: 55
Confidence: 7.5%
Identity: 16%
Fold: OB-fold
Superfamily: Hypothetical protein YgiW
Family: Hypothetical protein YgiW
Phyre2
| 15 |
|
PDB 1vf7 chain A
Region: 35 - 61
Aligned: 27
Modelled: 27
Confidence: 7.3%
Identity: 22%
Fold: HlyD-like secretion proteins
Superfamily: HlyD-like secretion proteins
Family: HlyD-like secretion proteins
Phyre2
| 16 |
|
PDB 2cp6 chain A domain 1
Region: 50 - 90
Aligned: 41
Modelled: 41
Confidence: 6.9%
Identity: 20%
Fold: SH3-like barrel
Superfamily: Cap-Gly domain
Family: Cap-Gly domain
Phyre2
| 17 |
|
PDB 1gtr chain A domain 1
Region: 41 - 89
Aligned: 49
Modelled: 49
Confidence: 6.8%
Identity: 8%
Fold: Ribosomal protein L25-like
Superfamily: Ribosomal protein L25-like
Family: Gln-tRNA synthetase (GlnRS), C-terminal (anticodon-binding) domain
Phyre2
| 18 |
|
PDB 2vv5 chain A domain 1
Region: 52 - 73
Aligned: 21
Modelled: 22
Confidence: 6.5%
Identity: 33%
Fold: Sm-like fold
Superfamily: Sm-like ribonucleoproteins
Family: Mechanosensitive channel protein MscS (YggB), middle domain
Phyre2