| 1 |
|
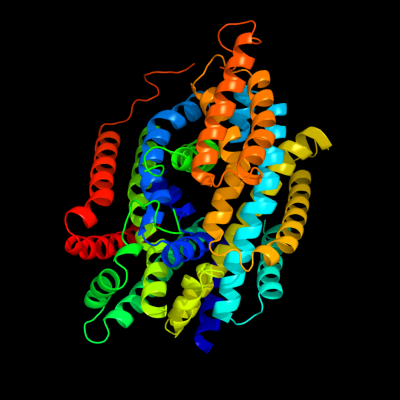
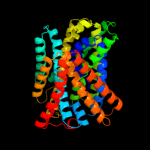
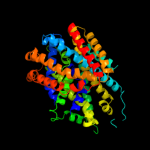
PDB 2xq2 chain A
Region: 2 - 556
Aligned: 521
Modelled: 521
Confidence: 100.0%
Identity: 25%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
Phyre2
| 2 |
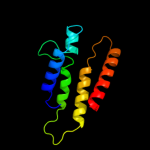
|
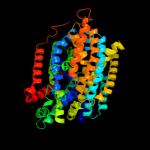
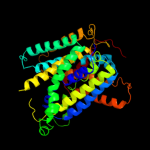
PDB 3dh4 chain A
Region: 23 - 523
Aligned: 489
Modelled: 474
Confidence: 100.0%
Identity: 26%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
Phyre2
| 3 |
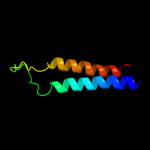
|
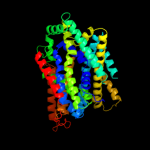
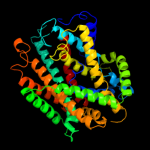
PDB 2jln chain A
Region: 25 - 497
Aligned: 440
Modelled: 440
Confidence: 99.4%
Identity: 10%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
| 4 |
|
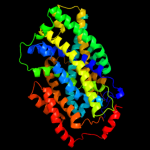
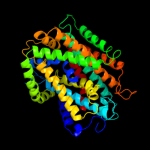
PDB 3gia chain A
Region: 39 - 494
Aligned: 429
Modelled: 429
Confidence: 99.1%
Identity: 9%
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized protein mj0609;
PDBTitle: crystal structure of apct transporter
Phyre2
| 5 |
|
PDB 3lrc chain C
Region: 40 - 487
Aligned: 402
Modelled: 402
Confidence: 98.2%
Identity: 8%
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
Phyre2
| 6 |
|
PDB 3hfx chain A
Region: 2 - 427
Aligned: 405
Modelled: 424
Confidence: 97.8%
Identity: 15%
PDB header:transport protein
Chain: A: PDB Molecule:l-carnitine/gamma-butyrobetaine antiporter;
PDBTitle: crystal structure of carnitine transporter
Phyre2
| 7 |
|
PDB 2w8a chain C
Region: 6 - 427
Aligned: 403
Modelled: 419
Confidence: 97.7%
Identity: 14%
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
Phyre2
| 8 |
|
PDB 2a65 chain A domain 1
Region: 49 - 524
Aligned: 437
Modelled: 437
Confidence: 85.7%
Identity: 12%
Fold: SNF-like
Superfamily: SNF-like
Family: SNF-like
Phyre2
| 9 |
|
PDB 3rlb chain A
Region: 375 - 481
Aligned: 98
Modelled: 107
Confidence: 17.0%
Identity: 17%
PDB header:thiamine-binding protein
Chain: A: PDB Molecule:thit;
PDBTitle: crystal structure at 2.0 a of the s-component for thiamin from an ecf-2 type abc transporter
Phyre2
| 10 |
|
PDB 2ky5 chain A
Region: 105 - 115
Aligned: 11
Modelled: 11
Confidence: 11.5%
Identity: 45%
PDB header:cell adhesion
Chain: A: PDB Molecule:platelet endothelial cell adhesion molecule;
PDBTitle: solution structure of the pecam-1 cytoplasmic tail with dpc
Phyre2
| 11 |
|
PDB 3mk7 chain F
Region: 3 - 66
Aligned: 64
Modelled: 64
Confidence: 8.1%
Identity: 16%
PDB header:oxidoreductase
Chain: F: PDB Molecule:cytochrome c oxidase, cbb3-type, subunit p;
PDBTitle: the structure of cbb3 cytochrome oxidase
Phyre2
| 12 |
|
PDB 3fxd chain D
Region: 560 - 569
Aligned: 10
Modelled: 10
Confidence: 7.5%
Identity: 70%
PDB header:unknown function
Chain: D: PDB Molecule:protein icmr;
PDBTitle: crystal structure of interacting domains of icmr and icmq
Phyre2
| 13 |
|
PDB 1fft chain B domain 2
Region: 532 - 571
Aligned: 40
Modelled: 40
Confidence: 7.4%
Identity: 13%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 14 |
|
PDB 3m7b chain A
Region: 429 - 565
Aligned: 135
Modelled: 137
Confidence: 7.1%
Identity: 13%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:tellurite resistance protein teha homolog;
PDBTitle: crystal structure of plant slac1 homolog teha
Phyre2
| 15 |
|
PDB 1w8x chain P
Region: 538 - 567
Aligned: 30
Modelled: 30
Confidence: 6.9%
Identity: 23%
PDB header:virus
Chain: P: PDB Molecule:protein p16;
PDBTitle: structural analysis of prd1
Phyre2
| 16 |
|
PDB 2fhz chain B domain 1
Region: 556 - 568
Aligned: 13
Modelled: 13
Confidence: 6.8%
Identity: 46%
Fold: Colicin D/E5 nuclease domain
Superfamily: Colicin D/E5 nuclease domain
Family: Colicin E5 nuclease domain
Phyre2
| 17 |
|
PDB 2axt chain I domain 1
Region: 538 - 562
Aligned: 24
Modelled: 25
Confidence: 6.5%
Identity: 29%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction center protein I, PsbI
Family: PsbI-like
Phyre2
| 18 |
|
PDB 1si2 chain A
Region: 103 - 114
Aligned: 12
Modelled: 12
Confidence: 6.2%
Identity: 17%
Fold: SH3-like barrel
Superfamily: PAZ domain
Family: PAZ domain
Phyre2
| 19 |
|
PDB 3o7x chain C
Region: 103 - 114
Aligned: 12
Modelled: 12
Confidence: 5.2%
Identity: 17%
PDB header:rna binding protein
Chain: C: PDB Molecule:piwi-like protein 2;
PDBTitle: crystal structure of human hili paz domain
Phyre2