| 1 |
|
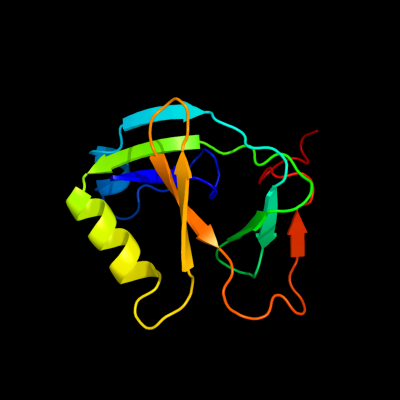
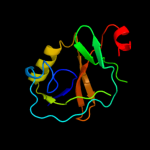
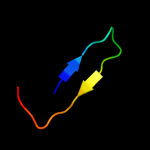
PDB 1xhs chain A
Region: 1 - 113
Aligned: 113
Modelled: 113
Confidence: 100.0%
Identity: 100%
Fold: Gamma-glutamyl cyclotransferase-like
Superfamily: Gamma-glutamyl cyclotransferase-like
Family: Gamma-glutamyl cyclotransferase-like
Phyre2
| 2 |
|
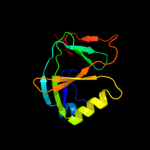
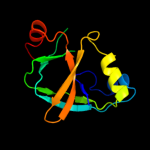
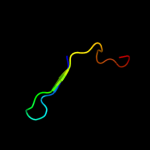
PDB 1v30 chain A
Region: 1 - 111
Aligned: 108
Modelled: 111
Confidence: 100.0%
Identity: 32%
Fold: Gamma-glutamyl cyclotransferase-like
Superfamily: Gamma-glutamyl cyclotransferase-like
Family: Gamma-glutamyl cyclotransferase-like
Phyre2
| 3 |
|
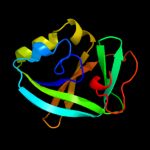
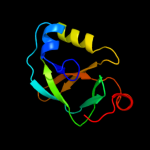
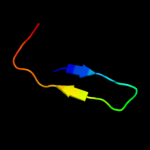
PDB 2qik chain A
Region: 3 - 109
Aligned: 99
Modelled: 103
Confidence: 100.0%
Identity: 29%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0131 protein ykqa;
PDBTitle: crystal structure of ykqa from bacillus subtilis. northeast2 structural genomics target sr631
Phyre2
| 4 |
|
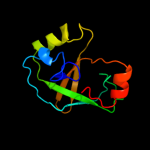
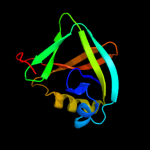
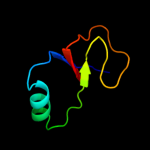
PDB 1vkb chain A
Region: 2 - 108
Aligned: 107
Modelled: 107
Confidence: 99.9%
Identity: 31%
Fold: Gamma-glutamyl cyclotransferase-like
Superfamily: Gamma-glutamyl cyclotransferase-like
Family: Gamma-glutamyl cyclotransferase-like
Phyre2
| 5 |
|
PDB 3jub chain A
Region: 1 - 104
Aligned: 104
Modelled: 104
Confidence: 99.9%
Identity: 27%
PDB header:transferase
Chain: A: PDB Molecule:aig2-like domain-containing protein 1;
PDBTitle: human gamma-glutamylamine cyclotransferase
Phyre2
| 6 |
|
PDB 2jqv chain A
Region: 2 - 108
Aligned: 102
Modelled: 107
Confidence: 99.9%
Identity: 25%
PDB header:structural genomics
Chain: A: PDB Molecule:aig2 protein-like;
PDBTitle: solution structure at3g28950.1 from arabidopsis thaliana
Phyre2
| 7 |
|
PDB 2g0q chain A
Region: 2 - 108
Aligned: 102
Modelled: 107
Confidence: 99.9%
Identity: 26%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:at5g39720.1 protein;
PDBTitle: solution structure of at5g39720.1 from arabidopsis thaliana
Phyre2
| 8 |
|
PDB 2rbh chain A
Region: 1 - 101
Aligned: 101
Modelled: 101
Confidence: 99.8%
Identity: 20%
PDB header:transferase
Chain: A: PDB Molecule:gamma-glutamyl cyclotransferase;
PDBTitle: gamma-glutamyl cyclotransferase
Phyre2
| 9 |
|
PDB 2o8r chain A domain 4
Region: 90 - 112
Aligned: 23
Modelled: 19
Confidence: 49.4%
Identity: 22%
Fold: Phospholipase D/nuclease
Superfamily: Phospholipase D/nuclease
Family: Polyphosphate kinase C-terminal domain
Phyre2
| 10 |
|
PDB 1xdp chain A domain 4
Region: 90 - 112
Aligned: 23
Modelled: 23
Confidence: 45.9%
Identity: 26%
Fold: Phospholipase D/nuclease
Superfamily: Phospholipase D/nuclease
Family: Polyphosphate kinase C-terminal domain
Phyre2
| 11 |
|
PDB 2o8r chain A
Region: 90 - 112
Aligned: 23
Modelled: 19
Confidence: 25.8%
Identity: 22%
PDB header:transferase
Chain: A: PDB Molecule:polyphosphate kinase;
PDBTitle: crystal structure of polyphosphate kinase from2 porphyromonas gingivalis
Phyre2
| 12 |
|
PDB 3eoi chain B
Region: 2 - 52
Aligned: 51
Modelled: 51
Confidence: 11.8%
Identity: 14%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:pilm;
PDBTitle: crystal structure of putative protein pilm from escherichia coli b7a
Phyre2
| 13 |
|
PDB 1hf2 chain A domain 1
Region: 2 - 10
Aligned: 9
Modelled: 9
Confidence: 8.2%
Identity: 22%
Fold: Single-stranded right-handed beta-helix
Superfamily: Cell-division inhibitor MinC, C-terminal domain
Family: Cell-division inhibitor MinC, C-terminal domain
Phyre2
| 14 |
|
PDB 1ynh chain A domain 1
Region: 1 - 7
Aligned: 7
Modelled: 7
Confidence: 7.9%
Identity: 71%
Fold: Pentein, beta/alpha-propeller
Superfamily: Pentein
Family: Succinylarginine dihydrolase-like
Phyre2
| 15 |
|
PDB 1xdo chain B
Region: 71 - 112
Aligned: 42
Modelled: 42
Confidence: 6.7%
Identity: 17%
PDB header:transferase
Chain: B: PDB Molecule:polyphosphate kinase;
PDBTitle: crystal structure of escherichia coli polyphosphate kinase
Phyre2
| 16 |
|
PDB 1hf2 chain A
Region: 2 - 10
Aligned: 9
Modelled: 9
Confidence: 5.4%
Identity: 22%
PDB header:cell division protein
Chain: A: PDB Molecule:septum site-determining protein minc;
PDBTitle: crystal structure of the bacterial cell-division inhibitor2 minc from t. maritima
Phyre2