| 1 |
|
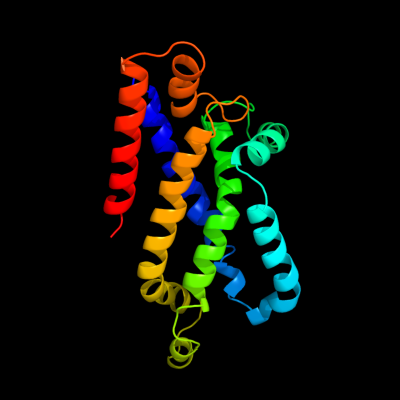
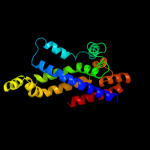
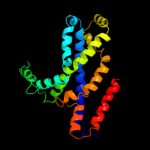
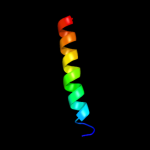
PDB 3dhw chain A domain 1
Region: 128 - 349
Aligned: 194
Modelled: 197
Confidence: 99.9%
Identity: 19%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 2 |
|
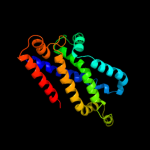
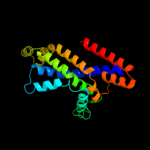
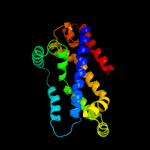
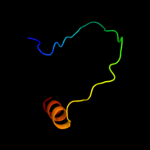
PDB 2onk chain C domain 1
Region: 127 - 351
Aligned: 206
Modelled: 208
Confidence: 99.8%
Identity: 17%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 3 |
|
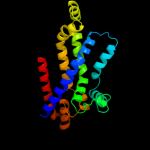
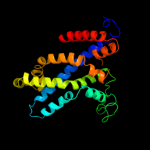
PDB 2onk chain C
Region: 127 - 351
Aligned: 206
Modelled: 208
Confidence: 99.8%
Identity: 17%
PDB header:membrane protein
Chain: C: PDB Molecule:molybdate/tungstate abc transporter, permease
PDBTitle: abc transporter modbc in complex with its binding protein2 moda
Phyre2
| 4 |
|
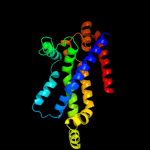
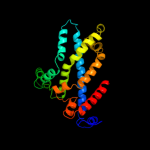
PDB 3d31 chain C domain 1
Region: 127 - 348
Aligned: 205
Modelled: 207
Confidence: 99.8%
Identity: 16%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 5 |
|
PDB 3d31 chain D
Region: 127 - 348
Aligned: 205
Modelled: 207
Confidence: 99.8%
Identity: 16%
PDB header:transport protein
Chain: D: PDB Molecule:sulfate/molybdate abc transporter, permease
PDBTitle: modbc from methanosarcina acetivorans
Phyre2
| 6 |
|
PDB 2r6g chain F domain 2
Region: 117 - 347
Aligned: 222
Modelled: 231
Confidence: 99.7%
Identity: 14%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 7 |
|
PDB 2r6g chain F
Region: 107 - 347
Aligned: 233
Modelled: 241
Confidence: 99.6%
Identity: 14%
PDB header:hydrolase/transport protein
Chain: F: PDB Molecule:maltose transport system permease protein malf;
PDBTitle: the crystal structure of the e. coli maltose transporter
Phyre2
| 8 |
|
PDB 3fh6 chain F
Region: 121 - 348
Aligned: 219
Modelled: 228
Confidence: 99.5%
Identity: 15%
PDB header:transport protein
Chain: F: PDB Molecule:maltose transport system permease protein malf;
PDBTitle: crystal structure of the resting state maltose transporter from e.2 coli
Phyre2
| 9 |
|
PDB 2r6g chain G domain 1
Region: 129 - 348
Aligned: 196
Modelled: 199
Confidence: 99.1%
Identity: 14%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 10 |
|
PDB 1wz4 chain A
Region: 353 - 360
Aligned: 8
Modelled: 8
Confidence: 21.8%
Identity: 50%
PDB header:gene regulation
Chain: A: PDB Molecule:major surface antigen;
PDBTitle: solution conformation of adr subtype hbv pre-s2 epitope
Phyre2
| 11 |
|
PDB 2cw1 chain A
Region: 250 - 273
Aligned: 22
Modelled: 24
Confidence: 18.4%
Identity: 41%
PDB header:de novo protein
Chain: A: PDB Molecule:sn4m;
PDBTitle: solution structure of the de novo-designed lambda cro fold2 protein
Phyre2
| 12 |
|
PDB 1cii chain A
Region: 112 - 169
Aligned: 55
Modelled: 58
Confidence: 16.1%
Identity: 11%
PDB header:transmembrane protein
Chain: A: PDB Molecule:colicin ia;
PDBTitle: colicin ia
Phyre2
| 13 |
|
PDB 1y66 chain D
Region: 82 - 105
Aligned: 24
Modelled: 24
Confidence: 13.6%
Identity: 21%
PDB header:de novo protein
Chain: D: PDB Molecule:engrailed homeodomain;
PDBTitle: dioxane contributes to the altered conformation and2 oligomerization state of a designed engrailed homeodomain3 variant
Phyre2
| 14 |
|
PDB 2ka1 chain A
Region: 137 - 161
Aligned: 25
Modelled: 25
Confidence: 11.3%
Identity: 32%
PDB header:membrane protein
Chain: A: PDB Molecule:bcl2/adenovirus e1b 19 kda protein-interacting
PDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles
Phyre2
| 15 |
|
PDB 2ka2 chain B
Region: 137 - 161
Aligned: 25
Modelled: 25
Confidence: 11.3%
Identity: 32%
PDB header:membrane protein
Chain: B: PDB Molecule:bcl2/adenovirus e1b 19 kda protein-interacting
PDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles with his173-ser172 intermonomer3 hydrogen bond restraints
Phyre2
| 16 |
|
PDB 2ka1 chain B
Region: 137 - 161
Aligned: 25
Modelled: 25
Confidence: 11.3%
Identity: 32%
PDB header:membrane protein
Chain: B: PDB Molecule:bcl2/adenovirus e1b 19 kda protein-interacting
PDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles
Phyre2
| 17 |
|
PDB 2ka2 chain A
Region: 137 - 161
Aligned: 25
Modelled: 25
Confidence: 11.3%
Identity: 32%
PDB header:membrane protein
Chain: A: PDB Molecule:bcl2/adenovirus e1b 19 kda protein-interacting
PDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles with his173-ser172 intermonomer3 hydrogen bond restraints
Phyre2
| 18 |
|
PDB 2hw2 chain A
Region: 31 - 93
Aligned: 38
Modelled: 38
Confidence: 9.7%
Identity: 21%
PDB header:transferase
Chain: A: PDB Molecule:rifampin adp-ribosyl transferase;
PDBTitle: crystal structure of rifampin adp-ribosyl transferase in2 complex with rifampin
Phyre2
| 19 |
|
PDB 2jwa chain A
Region: 135 - 164
Aligned: 30
Modelled: 30
Confidence: 9.1%
Identity: 17%
PDB header:transferase
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-2;
PDBTitle: erbb2 transmembrane segment dimer spatial structure
Phyre2
| 20 |
|
PDB 2j5d chain A
Region: 137 - 161
Aligned: 25
Modelled: 25
Confidence: 8.6%
Identity: 32%
PDB header:membrane protein
Chain: A: PDB Molecule:bcl2/adenovirus e1b 19 kda protein-interacting
PDBTitle: nmr structure of bnip3 transmembrane domain in lipid2 bicelles
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|
| 24 |
|