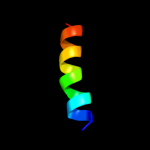| 1 |
|
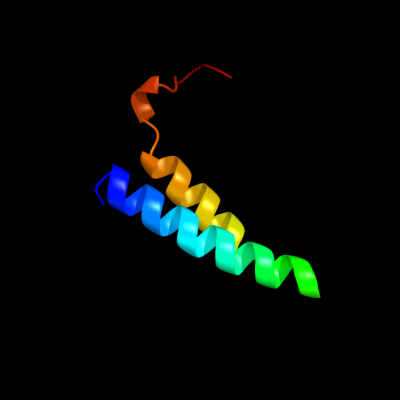
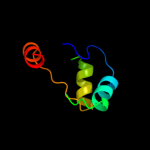
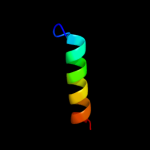
PDB 1sf9 chain A
Region: 1 - 51
Aligned: 51
Modelled: 51
Confidence: 73.9%
Identity: 37%
Fold: SH3-like barrel
Superfamily: Hypothetical protein YfhH
Family: Hypothetical protein YfhH
Phyre2
| 2 |
|
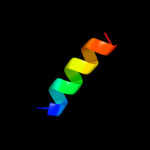
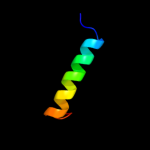
PDB 2yxy chain A
Region: 1 - 51
Aligned: 51
Modelled: 51
Confidence: 66.3%
Identity: 31%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical conserved protein, gk0453;
PDBTitle: crystarl structure of hypothetical conserved protein, gk0453
Phyre2
| 3 |
|
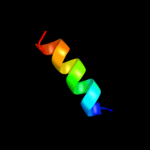
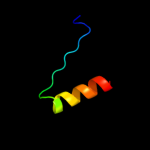
PDB 2kbz chain A
Region: 2 - 18
Aligned: 17
Modelled: 17
Confidence: 29.1%
Identity: 35%
PDB header:viral protein
Chain: A: PDB Molecule:15 protein (bacteriophage spp1 complete
PDBTitle: nmr structure of protein gp15 of bacteriophage spp1
Phyre2
| 4 |
|
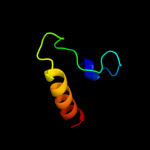
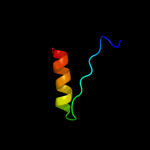
PDB 3h5j chain A
Region: 25 - 76
Aligned: 52
Modelled: 52
Confidence: 20.3%
Identity: 21%
PDB header:lyase
Chain: A: PDB Molecule:3-isopropylmalate dehydratase small subunit;
PDBTitle: leud_1-168 small subunit of isopropylmalate isomerase (rv2987c) from2 mycobacterium tuberculosis
Phyre2
| 5 |
|
PDB 1onv chain B
Region: 77 - 90
Aligned: 14
Modelled: 14
Confidence: 15.7%
Identity: 57%
PDB header:transcription
Chain: B: PDB Molecule:serine phosphatase fcp1a;
PDBTitle: nmr structure of a complex containing the tfiif subunit2 rap74 and the rnap ii ctd phosphatase fcp1
Phyre2
| 6 |
|
PDB 2e74 chain D domain 2
Region: 49 - 58
Aligned: 10
Modelled: 10
Confidence: 12.8%
Identity: 60%
Fold: Single transmembrane helix
Superfamily: ISP transmembrane anchor
Family: ISP transmembrane anchor
Phyre2
| 7 |
|
PDB 3q3w chain B
Region: 25 - 76
Aligned: 52
Modelled: 52
Confidence: 10.7%
Identity: 21%
PDB header:transferase
Chain: B: PDB Molecule:3-isopropylmalate dehydratase small subunit;
PDBTitle: isopropylmalate isomerase small subunit from campylobacter jejuni.
Phyre2
| 8 |
|
PDB 1qb2 chain A
Region: 1 - 29
Aligned: 29
Modelled: 29
Confidence: 10.5%
Identity: 24%
Fold: Signal peptide-binding domain
Superfamily: Signal peptide-binding domain
Family: Signal peptide-binding domain
Phyre2
| 9 |
|
PDB 1ifl chain A
Region: 24 - 42
Aligned: 18
Modelled: 19
Confidence: 9.8%
Identity: 50%
PDB header:virus
Chain: A: PDB Molecule:inovirus;
PDBTitle: molecular models and structural comparisons of native and2 mutant class i filamentous bacteriophages ff (fd, f1, m13),3 if1 and ike
Phyre2
| 10 |
|
PDB 2gbo chain B
Region: 7 - 23
Aligned: 17
Modelled: 15
Confidence: 8.7%
Identity: 41%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:upf0358 protein ef2458;
PDBTitle: protein of unknown function ef2458 from enterococcus faecalis
Phyre2
| 11 |
|
PDB 2gbo chain A domain 1
Region: 7 - 23
Aligned: 17
Modelled: 15
Confidence: 8.7%
Identity: 41%
Fold: Open three-helical up-and-down bundle
Superfamily: EF2458-like
Family: EF2458-like
Phyre2
| 12 |
|
PDB 1t0w chain A
Region: 41 - 51
Aligned: 11
Modelled: 11
Confidence: 7.9%
Identity: 45%
Fold: Knottins (small inhibitors, toxins, lectins)
Superfamily: Plant lectins/antimicrobial peptides
Family: Hevein-like agglutinin (lectin) domain
Phyre2
| 13 |
|
PDB 4a1a chain H
Region: 22 - 46
Aligned: 25
Modelled: 25
Confidence: 6.9%
Identity: 32%
PDB header:ribosome
Chain: H: PDB Molecule:60s ribosomal protein l10;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 3.
Phyre2
| 14 |
|
PDB 3t97 chain A
Region: 67 - 79
Aligned: 13
Modelled: 13
Confidence: 6.6%
Identity: 31%
PDB header:protein transport
Chain: A: PDB Molecule:nuclear pore glycoprotein p62;
PDBTitle: molecular architecture of the transport channel of the nuclear pore2 complex: nup62/nup54
Phyre2
| 15 |
|
PDB 1szh chain A
Region: 51 - 89
Aligned: 34
Modelled: 39
Confidence: 5.7%
Identity: 29%
Fold: Her-1
Superfamily: Her-1
Family: Her-1
Phyre2
| 16 |
|
PDB 2dw3 chain A
Region: 23 - 44
Aligned: 22
Modelled: 22
Confidence: 5.3%
Identity: 32%
PDB header:photosynthesis
Chain: A: PDB Molecule:intrinsic membrane protein pufx;
PDBTitle: solution structure of the rhodobacter sphaeroides pufx2 membrane protein
Phyre2
| 17 |
|
PDB 1gc5 chain A
Region: 1 - 39
Aligned: 39
Modelled: 39
Confidence: 5.2%
Identity: 13%
Fold: Ribokinase-like
Superfamily: Ribokinase-like
Family: ADP-specific Phosphofructokinase/Glucokinase
Phyre2
| 18 |
|
PDB 2pa2 chain A domain 1
Region: 22 - 47
Aligned: 26
Modelled: 26
Confidence: 5.2%
Identity: 35%
Fold: alpha/beta-Hammerhead
Superfamily: Ribosomal protein L16p/L10e
Family: Ribosomal protein L10e
Phyre2
| 19 |
|
PDB 1oft chain C
Region: 16 - 42
Aligned: 26
Modelled: 27
Confidence: 5.1%
Identity: 19%
PDB header:bacterial cell division inhibitor
Chain: C: PDB Molecule:hypothetical protein pa3008;
PDBTitle: crystal structure of sula from pseudomonas aeruginosa
Phyre2
| 20 |
|
PDB 1ofu chain X
Region: 16 - 42
Aligned: 26
Modelled: 27
Confidence: 5.0%
Identity: 19%
Fold: P-loop containing nucleoside triphosphate hydrolases
Superfamily: P-loop containing nucleoside triphosphate hydrolases
Family: Bacterial cell division inhibitor SulA
Phyre2