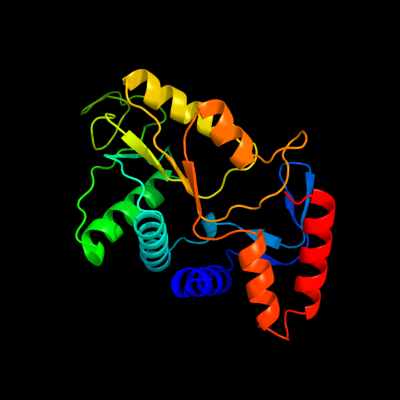
| 1 | d1lh0a_
|
|
 |
100.0 |
97 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
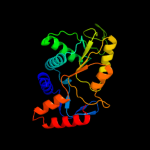
| 2 | c3n2lA_
|
|
 |
100.0 |
80 |
PDB header:transferase
Chain: A: PDB Molecule:orotate phosphoribosyltransferase;
PDBTitle: 2.1 angstrom resolution crystal structure of an orotate2 phosphoribosyltransferase (pyre) from vibrio cholerae o1 biovar eltor3 str. n16961
|
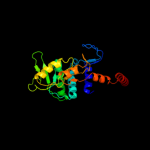
| 3 | c2przB_
|
|
 |
100.0 |
47 |
PDB header:transferase
Chain: B: PDB Molecule:orotate phosphoribosyltransferase 1;
PDBTitle: s. cerevisiae orotate phosphoribosyltransferase complexed2 with omp
|
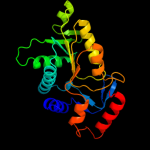
| 4 | c3mjdA_
|
|
 |
100.0 |
45 |
PDB header:transferase
Chain: A: PDB Molecule:orotate phosphoribosyltransferase;
PDBTitle: 1.9 angstrom crystal structure of orotate2 phosphoribosyltransferase (pyre) francisella tularensis.
|
| 5 | c2wnsB_
|
|
 |
100.0 |
28 |
PDB header:transferase
Chain: B: PDB Molecule:orotate phosphoribosyltransferase;
PDBTitle: human orotate phosphoribosyltransferase (oprtase) domain of2 uridine 5'-monophosphate synthase (umps) in complex with3 its substrate orotidine 5'-monophosphate (omp)
|
| 6 | c3qw4B_
|
|
 |
100.0 |
24 |
PDB header:transferase, lyase
Chain: B: PDB Molecule:ump synthase;
PDBTitle: structure of leishmania donovani ump synthase
|
| 7 | c3dezA_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:orotate phosphoribosyltransferase;
PDBTitle: crystal structure of orotate phosphoribosyltransferase from2 streptococcus mutans
|
| 8 | d2aeea1
|
|
 |
100.0 |
22 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 9 | c3m3hA_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:orotate phosphoribosyltransferase;
PDBTitle: 1.75 angstrom resolution crystal structure of an orotate2 phosphoribosyltransferase from bacillus anthracis str. 'ames3 ancestor'
|
| 10 | c2yzkC_
|
|
 |
100.0 |
27 |
PDB header:transferase
Chain: C: PDB Molecule:orotate phosphoribosyltransferase;
PDBTitle: crystal structure of orotate phosphoribosyltransferase from2 aeropyrum pernix
|
| 11 | d1mzva_
|
|
 |
100.0 |
20 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 12 | c2p1zA_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosyltransferase;
PDBTitle: crystal structure of phosphoribosyltransferase from corynebacterium2 diphtheriae
|
| 13 | d1qb7a_
|
|
 |
100.0 |
19 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 14 | d1y0ba1
|
|
 |
100.0 |
20 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 15 | d1o57a2
|
|
 |
99.9 |
21 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 16 | c2dy0A_
|
|
 |
99.9 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:adenine phosphoribosyltransferase;
PDBTitle: crystal structure of project jw0458 from escherichia coli
|
| 17 | c1o57A_
|
|
 |
99.9 |
19 |
PDB header:dna binding protein
Chain: A: PDB Molecule:pur operon repressor;
PDBTitle: crystal structure of the purine operon repressor of2 bacillus subtilis
|
| 18 | d1zn7a1
|
|
 |
99.9 |
19 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 19 | d1l1qa_
|
|
 |
99.9 |
18 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 20 | d1g2qa_
|
|
 |
99.9 |
23 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 21 | d1vcha1 |
|
not modelled |
99.8 |
23 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 22 | d1vdma1 |
|
not modelled |
99.8 |
19 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 23 | d2igba1 |
|
not modelled |
99.8 |
19 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 24 | d1ufra_ |
|
not modelled |
99.7 |
20 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 25 | d1hgxa_ |
|
not modelled |
99.7 |
15 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 26 | d1w30a_ |
|
not modelled |
99.7 |
17 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 27 | c3o7mD_ |
|
not modelled |
99.7 |
11 |
PDB header:transferase
Chain: D: PDB Molecule:hypoxanthine phosphoribosyltransferase;
PDBTitle: 1.98 angstrom resolution crystal structure of a hypoxanthine-guanine2 phosphoribosyltransferase (hpt-2) from bacillus anthracis str. 'ames3 ancestor'
|
| 28 | d1a3ca_ |
|
not modelled |
99.7 |
17 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 29 | d1g9sa_ |
|
not modelled |
99.7 |
13 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 30 | c2ywtA_ |
|
not modelled |
99.7 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:hypoxanthine-guanine phosphoribosyltransferase;
PDBTitle: crystal structure of hypoxanthine-guanine2 phosphoribosyltransferase with gmp from thermus3 thermophilus hb8
|
| 31 | d1j7ja_ |
|
not modelled |
99.7 |
12 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 32 | d1z7ga1 |
|
not modelled |
99.7 |
12 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 33 | d1pzma_ |
|
not modelled |
99.7 |
15 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 34 | d1tc1a_ |
|
not modelled |
99.7 |
13 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 35 | c3kb8A_ |
|
not modelled |
99.7 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:hypoxanthine phosphoribosyltransferase;
PDBTitle: 2.09 angstrom resolution structure of a hypoxanthine-guanine2 phosphoribosyltransferase (hpt-1) from bacillus anthracis str. 'ames3 ancestor' in complex with gmp
|
| 36 | c1pzmB_ |
|
not modelled |
99.6 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:hypoxanthine-guanine phosphoribosyltransferase;
PDBTitle: crystal structure of hgprt-ase from leishmania tarentolae in complex2 with gmp
|
| 37 | d1yfza1 |
|
not modelled |
99.6 |
18 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 38 | c1yfzA_ |
|
not modelled |
99.6 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:hypoxanthine-guanine phosphoribosyltransferase;
PDBTitle: novel imp binding in feedback inhibition of hypoxanthine-guanine2 phosphoribosyltransferase from thermoanaerobacter tengcongensis
|
| 39 | d1p17b_ |
|
not modelled |
99.6 |
13 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 40 | c2jkzB_ |
|
not modelled |
99.6 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:hypoxanthine-guanine phosphoribosyltransferase;
PDBTitle: saccharomyces cerevisiae hypoxanthine-guanine2 phosphoribosyltransferase in complex with gmp (guanosine 5'3 -monophosphate) (orthorhombic crystal form)
|
| 41 | d1ecfa1 |
|
not modelled |
99.5 |
17 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 42 | d1nula_ |
|
not modelled |
99.5 |
13 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 43 | d1gph11 |
|
not modelled |
99.5 |
25 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 44 | c2jbhA_ |
|
not modelled |
99.5 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosyltransferase domain-containing protein 1;
PDBTitle: human phosphoribosyl transferase domain containing 1
|
| 45 | c3lpnB_ |
|
not modelled |
99.5 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:ribose-phosphate pyrophosphokinase;
PDBTitle: crystal structure of the phosphoribosylpyrophosphate (prpp) synthetase2 from thermoplasma volcanium in complex with an atp analog (ampcpp).
|
| 46 | c1ecjB_ |
|
not modelled |
99.4 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:glutamine phosphoribosylpyrophosphate
PDBTitle: escherichia coli glutamine phosphoribosylpyrophosphate2 (prpp) amidotransferase complexed with 2 amp per tetramer
|
| 47 | d1wd5a_ |
|
not modelled |
99.4 |
24 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 48 | c3efhB_ |
|
not modelled |
99.4 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:ribose-phosphate pyrophosphokinase 1;
PDBTitle: crystal structure of human phosphoribosyl pyrophosphate2 synthetase 1
|
| 49 | d1fsga_ |
|
not modelled |
99.4 |
13 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 50 | d1cjba_ |
|
not modelled |
99.4 |
11 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 51 | c1gph1_ |
|
not modelled |
99.4 |
26 |
PDB header:transferase(glutamine amidotransferase)
Chain: 1: PDB Molecule:glutamine phosphoribosyl-pyrophosphate amidotransferase;
PDBTitle: structure of the allosteric regulatory enzyme of purine biosynthesis
|
| 52 | c1dkrB_ |
|
not modelled |
99.3 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:phosphoribosyl pyrophosphate synthetase;
PDBTitle: crystal structures of bacillus subtilis phosphoribosylpyrophosphate2 synthetase: molecular basis of allosteric inhibition and activation.
|
| 53 | c2c4kD_ |
|
not modelled |
99.2 |
17 |
PDB header:regulatory protein
Chain: D: PDB Molecule:phosphoribosyl pyrophosphate synthetase-
PDBTitle: crystal structure of human phosphoribosylpyrophosphate2 synthetase-associated protein 39 (pap39)
|
| 54 | d1dkua2 |
|
not modelled |
99.2 |
24 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosylpyrophosphate synthetase-like |
| 55 | d1u9ya2 |
|
not modelled |
99.2 |
21 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosylpyrophosphate synthetase-like |
| 56 | d2c4ka2 |
|
not modelled |
99.2 |
22 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosylpyrophosphate synthetase-like |
| 57 | c1u9yD_ |
|
not modelled |
99.1 |
21 |
PDB header:transferase
Chain: D: PDB Molecule:ribose-phosphate pyrophosphokinase;
PDBTitle: crystal structure of phosphoribosyl diphosphate synthase2 from methanocaldococcus jannaschii
|
| 58 | c3dahB_ |
|
not modelled |
98.9 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:ribose-phosphate pyrophosphokinase;
PDBTitle: 2.3 a crystal structure of ribose-phosphate pyrophosphokinase from2 burkholderia pseudomallei
|
| 59 | d1dqna_ |
|
not modelled |
98.3 |
17 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 60 | c2ehjA_ |
|
not modelled |
98.3 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:uracil phosphoribosyltransferase;
PDBTitle: structure of uracil phosphoribosyl transferase
|
| 61 | d1o5oa_ |
|
not modelled |
98.2 |
15 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 62 | c2e55D_ |
|
not modelled |
98.2 |
9 |
PDB header:transferase
Chain: D: PDB Molecule:uracil phosphoribosyltransferase;
PDBTitle: structure of aq2163 protein from aquifex aeolicus
|
| 63 | d1i5ea_ |
|
not modelled |
98.1 |
11 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 64 | d1xtta1 |
|
not modelled |
97.7 |
9 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 65 | d1bd3a_ |
|
not modelled |
97.7 |
13 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 66 | c3dmpD_ |
|
not modelled |
97.6 |
10 |
PDB header:transferase
Chain: D: PDB Molecule:uracil phosphoribosyltransferase;
PDBTitle: 2.6 a crystal structure of uracil phosphoribosyltransferase2 from burkholderia pseudomallei
|
| 67 | d1v9sa1 |
|
not modelled |
97.4 |
14 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 68 | d1g57a_ |
|
not modelled |
91.5 |
25 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:3,4-dihydroxy-2-butanone 4-phosphate synthase, DHBP synthase, RibB |
| 69 | d1tksa_ |
|
not modelled |
90.8 |
14 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:3,4-dihydroxy-2-butanone 4-phosphate synthase, DHBP synthase, RibB |
| 70 | d1snna_ |
|
not modelled |
89.5 |
8 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:3,4-dihydroxy-2-butanone 4-phosphate synthase, DHBP synthase, RibB |
| 71 | c3mioA_ |
|
not modelled |
89.3 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:3,4-dihydroxy-2-butanone 4-phosphate synthase;
PDBTitle: crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase2 domain from mycobacterium tuberculosis at ph 6.00
|
| 72 | d1k4ia_ |
|
not modelled |
83.5 |
20 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:3,4-dihydroxy-2-butanone 4-phosphate synthase, DHBP synthase, RibB |
| 73 | d1s8na_ |
|
not modelled |
79.4 |
18 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 74 | d1u9ya1 |
|
not modelled |
69.7 |
11 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosylpyrophosphate synthetase-like |
| 75 | c2vspA_ |
|
not modelled |
62.3 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:pdz domain-containing protein 1;
PDBTitle: crystal structure of the fourth pdz domain of pdz domain-2 containing protein 1
|
| 76 | d1vaea_ |
|
not modelled |
60.4 |
12 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 77 | c3l4fD_ |
|
not modelled |
59.9 |
18 |
PDB header:signaling protein/protein binding
Chain: D: PDB Molecule:sh3 and multiple ankyrin repeat domains protein
PDBTitle: crystal structure of betapix coiled-coil domain and shank2 pdz complex
|
| 78 | c2vsvB_ |
|
not modelled |
57.2 |
12 |
PDB header:protein-binding
Chain: B: PDB Molecule:rhophilin-2;
PDBTitle: crystal structure of the pdz domain of human rhophilin-2
|
| 79 | d2c4ka1 |
|
not modelled |
57.1 |
11 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosylpyrophosphate synthetase-like |
| 80 | d1m5za_ |
|
not modelled |
54.8 |
23 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 81 | c3eggC_ |
|
not modelled |
54.4 |
10 |
PDB header:hydrolase
Chain: C: PDB Molecule:spinophilin;
PDBTitle: crystal structure of a complex between protein phosphatase 1 alpha2 (pp1) and the pp1 binding and pdz domains of spinophilin
|
| 82 | c2d90A_ |
|
not modelled |
54.1 |
15 |
PDB header:protein binding
Chain: A: PDB Molecule:pdz domain containing protein 1;
PDBTitle: solution structure of the third pdz domain of pdz domain2 containing protein 1
|
| 83 | c3r2uC_ |
|
not modelled |
52.6 |
18 |
PDB header:hydrolase
Chain: C: PDB Molecule:metallo-beta-lactamase family protein;
PDBTitle: 2.1 angstrom resolution crystal structure of metallo-beta-lactamase2 from staphylococcus aureus subsp. aureus col
|
| 84 | c2jxoA_ |
|
not modelled |
52.6 |
23 |
PDB header:protein binding
Chain: A: PDB Molecule:ezrin-radixin-moesin-binding phosphoprotein 50;
PDBTitle: structure of the second pdz domain of nherf-1
|
| 85 | c3mmnA_ |
|
not modelled |
51.8 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:histidine kinase homolog;
PDBTitle: crystal structure of the receiver domain of the histidine kinase cki12 from arabidopsis thaliana complexed with mg2+
|
| 86 | c1a2oB_ |
|
not modelled |
51.5 |
20 |
PDB header:bacterial chemotaxis
Chain: B: PDB Molecule:cheb methylesterase;
PDBTitle: structural basis for methylesterase cheb regulation by a2 phosphorylation-activated domain
|
| 87 | d1q3oa_ |
|
not modelled |
51.3 |
18 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 88 | c2o2tB_ |
|
not modelled |
50.8 |
10 |
PDB header:structural protein
Chain: B: PDB Molecule:multiple pdz domain protein;
PDBTitle: the crystal structure of the 1st pdz domain of mpdz
|
| 89 | c3diwB_ |
|
not modelled |
48.2 |
10 |
PDB header:signaling protein/cell adhesion
Chain: B: PDB Molecule:tax1-binding protein 3;
PDBTitle: c-terminal beta-catenin bound tip-1 structure
|
| 90 | c2kjdA_ |
|
not modelled |
47.5 |
23 |
PDB header:signaling protein
Chain: A: PDB Molecule:sodium/hydrogen exchange regulatory cofactor nhe-
PDBTitle: solution structure of extended pdz2 domain from nherf1 (150-2 270)
|
| 91 | d1ihja_ |
|
not modelled |
47.5 |
6 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 92 | c3qglD_ |
|
not modelled |
47.2 |
20 |
PDB header:protein binding
Chain: D: PDB Molecule:sorting nexin-27;
PDBTitle: crystal structure of pdz domain of sorting nexin 27 (snx27) in complex2 with the eseskv peptide corresponding to the c-terminal tail of girk3
|
| 93 | c1r6jA_ |
|
not modelled |
47.1 |
10 |
PDB header:membrane protein
Chain: A: PDB Molecule:syntenin 1;
PDBTitle: ultrahigh resolution crystal structure of syntenin pdz2
|
| 94 | d1r6ja_ |
|
not modelled |
47.1 |
10 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 95 | c1nteA_ |
|
not modelled |
47.1 |
10 |
PDB header:signaling protein
Chain: A: PDB Molecule:syntenin 1;
PDBTitle: crystal structure analysis of the second pdz domain of2 syntenin
|
| 96 | c1j6uA_ |
|
not modelled |
46.6 |
11 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate-alanine ligase murc;
PDBTitle: crystal structure of udp-n-acetylmuramate-alanine ligase2 murc (tm0231) from thermotoga maritima at 2.3 a resolution
|
| 97 | c2eeiA_ |
|
not modelled |
46.2 |
15 |
PDB header:metal binding protein
Chain: A: PDB Molecule:pdz domain-containing protein 1;
PDBTitle: solution structure of second pdz domain of pdz domain2 containing protein 1
|
| 98 | c1obyA_ |
|
not modelled |
46.0 |
10 |
PDB header:cell adhesion
Chain: A: PDB Molecule:syntenin 1;
PDBTitle: crystal structure of the complex of pdz2 of syntenin with2 a syndecan-4 peptide.
|
| 99 | d1dcfa_ |
|
not modelled |
44.9 |
15 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:Receiver domain of the ethylene receptor |
| 100 | d1vi2a1 |
|
not modelled |
44.2 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 101 | d1dkua1 |
|
not modelled |
44.2 |
11 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosylpyrophosphate synthetase-like |
| 102 | c2kv8A_ |
|
not modelled |
43.8 |
13 |
PDB header:signaling protein
Chain: A: PDB Molecule:regulator of g-protein signaling 12;
PDBTitle: solution structure ofrgs12 pdz domain
|
| 103 | c2he4A_ |
|
not modelled |
43.7 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:na(+)/h(+) exchange regulatory cofactor nhe-rf2;
PDBTitle: the crystal structure of the second pdz domain of human2 nherf-2 (slc9a3r2) interacting with a mode 1 pdz binding3 motif
|
| 104 | c2z17A_ |
|
not modelled |
43.5 |
19 |
PDB header:protein binding
Chain: A: PDB Molecule:pleckstrin homology sec7 and coiled-coil domains-
PDBTitle: crystal sturcture of pdz domain from human pleckstrin2 homology, sec7
|
| 105 | d1v6ba_ |
|
not modelled |
43.0 |
18 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 106 | d2fe5a1 |
|
not modelled |
40.6 |
8 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 107 | d1g9oa_ |
|
not modelled |
40.6 |
13 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 108 | d1y7na1 |
|
not modelled |
40.2 |
10 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 109 | c3k1rA_ |
|
not modelled |
40.0 |
10 |
PDB header:structural protein
Chain: A: PDB Molecule:harmonin;
PDBTitle: structure of harmonin npdz1 in complex with the sam-pbm of2 sans
|
| 110 | c2edzA_ |
|
not modelled |
40.0 |
15 |
PDB header:signaling protein
Chain: A: PDB Molecule:pdz domain-containing protein 1;
PDBTitle: solution structures of the pdz domain of mus musculus pdz2 domain-containing protein 1
|
| 111 | c2krgA_ |
|
not modelled |
39.9 |
23 |
PDB header:signaling protein
Chain: A: PDB Molecule:na(+)/h(+) exchange regulatory cofactor nhe-rf1;
PDBTitle: solution structure of human sodium/ hydrogen exchange2 regulatory factor 1(150-358)
|
| 112 | c2pkuA_ |
|
not modelled |
39.9 |
15 |
PDB header:protein binding
Chain: A: PDB Molecule:prkca-binding protein;
PDBTitle: solution structure of pick1 pdz in complex with the2 carboxyl tail peptide of glur2
|
| 113 | d1u3ba2 |
|
not modelled |
39.8 |
11 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 114 | c2dmzA_ |
|
not modelled |
39.7 |
20 |
PDB header:protein binding
Chain: A: PDB Molecule:inad-like protein;
PDBTitle: solution structure of the third pdz domain of human inad-2 like protein
|
| 115 | c1obyB_ |
|
not modelled |
39.3 |
10 |
PDB header:cell adhesion
Chain: B: PDB Molecule:syntenin 1;
PDBTitle: crystal structure of the complex of pdz2 of syntenin with2 a syndecan-4 peptide.
|
| 116 | d1obxa_ |
|
not modelled |
39.3 |
10 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 117 | c1obxA_ |
|
not modelled |
39.3 |
10 |
PDB header:cell adhesion
Chain: A: PDB Molecule:syntenin 1;
PDBTitle: crystal structure of the complex of pdz2 of syntenin with2 an interleukin 5 receptor alpha peptide.
|
| 118 | d2j01f1 |
|
not modelled |
38.9 |
13 |
Fold:Ribosomal protein L4
Superfamily:Ribosomal protein L4
Family:Ribosomal protein L4 |
| 119 | d1uita_ |
|
not modelled |
38.1 |
10 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 120 | d1wjla_ |
|
not modelled |
38.1 |
21 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |