| 1 |
|
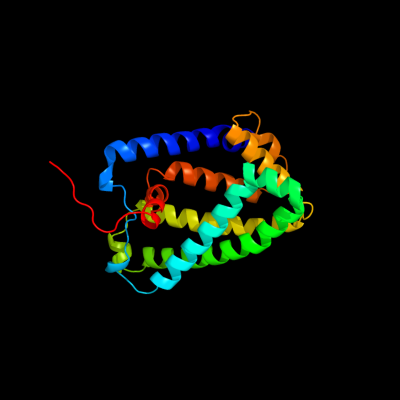
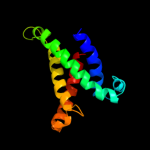
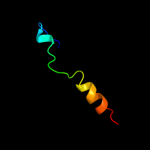
PDB 1y5i chain C domain 1
Region: 3 - 225
Aligned: 215
Modelled: 223
Confidence: 100.0%
Identity: 72%
Fold: Heme-binding four-helical bundle
Superfamily: Respiratory nitrate reductase 1 gamma chain
Family: Respiratory nitrate reductase 1 gamma chain
Phyre2
| 2 |
|
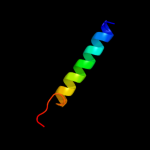
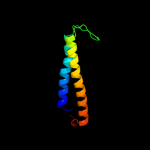
PDB 2knc chain A
Region: 92 - 127
Aligned: 36
Modelled: 36
Confidence: 75.6%
Identity: 22%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 3 |
|
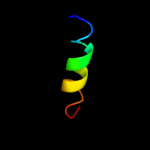
PDB 2klu chain A
Region: 92 - 123
Aligned: 32
Modelled: 32
Confidence: 71.7%
Identity: 34%
PDB header:immune system, membrane protein
Chain: A: PDB Molecule:t-cell surface glycoprotein cd4;
PDBTitle: nmr structure of the transmembrane and cytoplasmic domains2 of human cd4
Phyre2
| 4 |
|
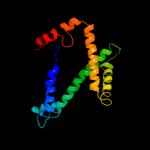
PDB 1kqf chain C
Region: 55 - 200
Aligned: 123
Modelled: 123
Confidence: 57.7%
Identity: 15%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Formate dehydrogenase N, cytochrome (gamma) subunit
Phyre2
| 5 |
|
PDB 2k1a chain A
Region: 92 - 120
Aligned: 29
Modelled: 29
Confidence: 39.1%
Identity: 24%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
Phyre2
| 6 |
|
PDB 3kdp chain H
Region: 97 - 113
Aligned: 17
Modelled: 17
Confidence: 38.9%
Identity: 24%
PDB header:hydrolase
Chain: H: PDB Molecule:na+/k+ atpase gamma subunit transcript variant a;
PDBTitle: crystal structure of the sodium-potassium pump
Phyre2
| 7 |
|
PDB 3kdp chain G
Region: 97 - 113
Aligned: 17
Modelled: 17
Confidence: 38.9%
Identity: 24%
PDB header:hydrolase
Chain: G: PDB Molecule:na+/k+ atpase gamma subunit transcript variant a;
PDBTitle: crystal structure of the sodium-potassium pump
Phyre2
| 8 |
|
PDB 2w8a chain C
Region: 5 - 168
Aligned: 156
Modelled: 163
Confidence: 18.2%
Identity: 11%
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
Phyre2
| 9 |
|
PDB 2jp3 chain A
Region: 87 - 124
Aligned: 38
Modelled: 38
Confidence: 13.0%
Identity: 18%
PDB header:transcription
Chain: A: PDB Molecule:fxyd domain-containing ion transport regulator 4;
PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
Phyre2
| 10 |
|
PDB 2k1l chain B
Region: 87 - 113
Aligned: 27
Modelled: 27
Confidence: 11.9%
Identity: 26%
PDB header:signaling protein
Chain: B: PDB Molecule:ephrin type-a receptor 1;
PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 6.3
Phyre2
| 11 |
|
PDB 2k1k chain B
Region: 87 - 113
Aligned: 27
Modelled: 27
Confidence: 11.9%
Identity: 26%
PDB header:signaling protein
Chain: B: PDB Molecule:ephrin type-a receptor 1;
PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 4.3
Phyre2
| 12 |
|
PDB 2k1l chain A
Region: 87 - 113
Aligned: 27
Modelled: 27
Confidence: 11.9%
Identity: 26%
PDB header:signaling protein
Chain: A: PDB Molecule:ephrin type-a receptor 1;
PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 6.3
Phyre2
| 13 |
|
PDB 2k1k chain A
Region: 87 - 113
Aligned: 27
Modelled: 27
Confidence: 11.9%
Identity: 26%
PDB header:signaling protein
Chain: A: PDB Molecule:ephrin type-a receptor 1;
PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 4.3
Phyre2
| 14 |
|
PDB 2k9y chain B
Region: 83 - 113
Aligned: 31
Modelled: 31
Confidence: 10.2%
Identity: 26%
PDB header:transferase
Chain: B: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
Phyre2
| 15 |
|
PDB 2jo1 chain A
Region: 87 - 117
Aligned: 31
Modelled: 31
Confidence: 9.8%
Identity: 26%
PDB header:hydrolase regulator
Chain: A: PDB Molecule:phospholemman;
PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
Phyre2
| 16 |
|
PDB 2k9y chain A
Region: 83 - 113
Aligned: 31
Modelled: 31
Confidence: 7.5%
Identity: 26%
PDB header:transferase
Chain: A: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
Phyre2
| 17 |
|
PDB 2dyo chain B
Region: 109 - 120
Aligned: 12
Modelled: 12
Confidence: 6.7%
Identity: 50%
PDB header:protein turnover/protein turnover
Chain: B: PDB Molecule:autophagy protein 16;
PDBTitle: the crystal structure of saccharomyces cerevisiae atg5-2 atg16(1-57) complex
Phyre2
| 18 |
|
PDB 1ar1 chain A
Region: 44 - 123
Aligned: 80
Modelled: 80
Confidence: 5.6%
Identity: 16%
Fold: Cytochrome c oxidase subunit I-like
Superfamily: Cytochrome c oxidase subunit I-like
Family: Cytochrome c oxidase subunit I-like
Phyre2