| 1 |
|
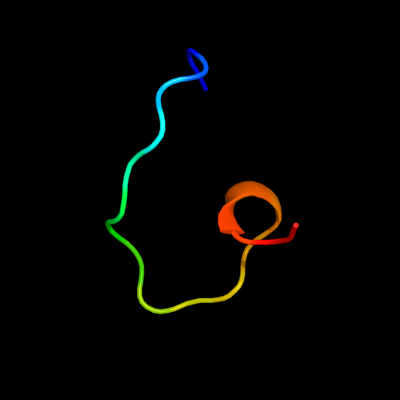
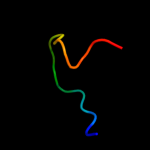
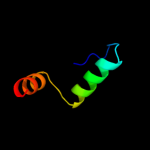
PDB 2yug chain A
Region: 21 - 38
Aligned: 18
Modelled: 18
Confidence: 14.3%
Identity: 22%
PDB header:gene regulation
Chain: A: PDB Molecule:protein frg1;
PDBTitle: solution structure of mouse frg1 protein
Phyre2
| 2 |
|
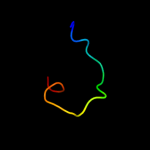
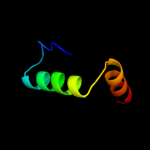
PDB 1dfc chain A domain 3
Region: 24 - 38
Aligned: 15
Modelled: 15
Confidence: 12.9%
Identity: 20%
Fold: beta-Trefoil
Superfamily: Actin-crosslinking proteins
Family: Fascin
Phyre2
| 3 |
|
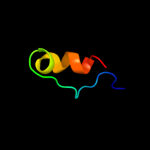
PDB 1ecf chain A domain 1
Region: 10 - 19
Aligned: 10
Modelled: 10
Confidence: 11.9%
Identity: 40%
Fold: PRTase-like
Superfamily: PRTase-like
Family: Phosphoribosyltransferases (PRTases)
Phyre2
| 4 |
|
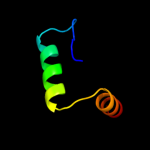
PDB 1ecj chain B
Region: 10 - 19
Aligned: 10
Modelled: 10
Confidence: 11.1%
Identity: 40%
PDB header:transferase
Chain: B: PDB Molecule:glutamine phosphoribosylpyrophosphate
PDBTitle: escherichia coli glutamine phosphoribosylpyrophosphate2 (prpp) amidotransferase complexed with 2 amp per tetramer
Phyre2
| 5 |
|
PDB 3soo chain B
Region: 24 - 35
Aligned: 12
Modelled: 12
Confidence: 10.9%
Identity: 33%
PDB header:rna binding protein
Chain: B: PDB Molecule:line-1 type transposase domain-containing protein 1;
PDBTitle: crystal structure of a line-1 type transposase domain-containing2 protein 1 (l1td1) from homo sapiens at 2.73 a resolution
Phyre2
| 6 |
|
PDB 1cgm chain E
Region: 14 - 70
Aligned: 53
Modelled: 57
Confidence: 10.7%
Identity: 11%
Fold: Four-helical up-and-down bundle
Superfamily: TMV-like viral coat proteins
Family: TMV-like viral coat proteins
Phyre2
| 7 |
|
PDB 1gph chain 1 domain 1
Region: 10 - 19
Aligned: 10
Modelled: 10
Confidence: 9.8%
Identity: 50%
Fold: PRTase-like
Superfamily: PRTase-like
Family: Phosphoribosyltransferases (PRTases)
Phyre2
| 8 |
|
PDB 1c0m chain A domain 1
Region: 22 - 36
Aligned: 15
Modelled: 15
Confidence: 9.6%
Identity: 33%
Fold: SH3-like barrel
Superfamily: DNA-binding domain of retroviral integrase
Family: DNA-binding domain of retroviral integrase
Phyre2
| 9 |
|
PDB 1gph chain 1
Region: 10 - 19
Aligned: 10
Modelled: 10
Confidence: 9.5%
Identity: 50%
PDB header:transferase(glutamine amidotransferase)
Chain: 1: PDB Molecule:glutamine phosphoribosyl-pyrophosphate amidotransferase;
PDBTitle: structure of the allosteric regulatory enzyme of purine biosynthesis
Phyre2
| 10 |
|
PDB 2vv5 chain A domain 1
Region: 14 - 34
Aligned: 21
Modelled: 21
Confidence: 9.4%
Identity: 10%
Fold: Sm-like fold
Superfamily: Sm-like ribonucleoproteins
Family: Mechanosensitive channel protein MscS (YggB), middle domain
Phyre2
| 11 |
|
PDB 1dfc chain A domain 1
Region: 23 - 38
Aligned: 16
Modelled: 16
Confidence: 8.5%
Identity: 19%
Fold: beta-Trefoil
Superfamily: Actin-crosslinking proteins
Family: Fascin
Phyre2
| 12 |
|
PDB 1dfc chain A domain 2
Region: 21 - 38
Aligned: 18
Modelled: 18
Confidence: 7.5%
Identity: 22%
Fold: beta-Trefoil
Superfamily: Actin-crosslinking proteins
Family: Fascin
Phyre2
| 13 |
|
PDB 3aqb chain C
Region: 15 - 25
Aligned: 11
Modelled: 11
Confidence: 6.2%
Identity: 45%
PDB header:transferase
Chain: C: PDB Molecule:component a of hexaprenyl diphosphate synthase;
PDBTitle: m. luteus b-p 26 heterodimeric hexaprenyl diphosphate synthase in2 complex with magnesium
Phyre2
| 14 |
|
PDB 2w0c chain C
Region: 14 - 38
Aligned: 22
Modelled: 25
Confidence: 5.8%
Identity: 18%
PDB header:virus
Chain: C: PDB Molecule:major capsid protein p2;
PDBTitle: x-ray structure of the entire lipid-containing2 bacteriophage pm2
Phyre2
| 15 |
|
PDB 1qop chain A
Region: 16 - 50
Aligned: 35
Modelled: 35
Confidence: 5.6%
Identity: 14%
Fold: TIM beta/alpha-barrel
Superfamily: Ribulose-phoshate binding barrel
Family: Tryptophan biosynthesis enzymes
Phyre2
| 16 |
|
PDB 3nav chain B
Region: 16 - 50
Aligned: 35
Modelled: 35
Confidence: 5.6%
Identity: 17%
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
Phyre2
| 17 |
|
PDB 2ekc chain A
Region: 16 - 50
Aligned: 35
Modelled: 35
Confidence: 5.5%
Identity: 23%
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
Phyre2