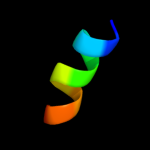| 1 |
|
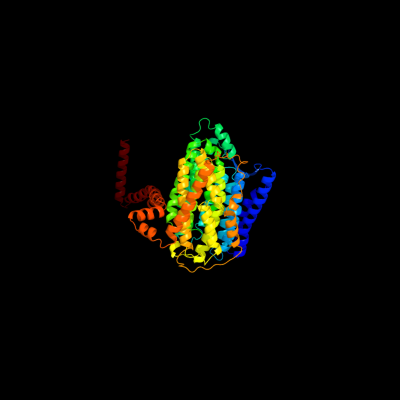
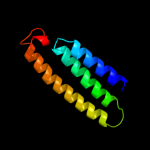
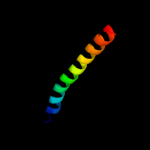
PDB 3rko chain L
Region: 1 - 612
Aligned: 612
Modelled: 612
Confidence: 100.0%
Identity: 100%
PDB header:oxidoreductase
Chain: L: PDB Molecule:nadh-quinone oxidoreductase subunit l;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 2 |
|
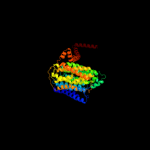
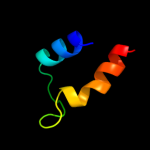
PDB 3rko chain M
Region: 3 - 487
Aligned: 479
Modelled: 484
Confidence: 100.0%
Identity: 20%
PDB header:oxidoreductase
Chain: M: PDB Molecule:nadh-quinone oxidoreductase subunit m;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 3 |
|
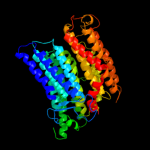
PDB 3rko chain N
Region: 1 - 483
Aligned: 459
Modelled: 467
Confidence: 100.0%
Identity: 19%
PDB header:oxidoreductase
Chain: N: PDB Molecule:nadh-quinone oxidoreductase subunit n;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 4 |
|
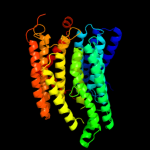
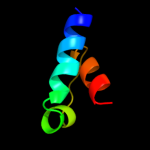
PDB 3rko chain K
Region: 116 - 200
Aligned: 85
Modelled: 85
Confidence: 83.0%
Identity: 22%
PDB header:oxidoreductase
Chain: K: PDB Molecule:nadh-quinone oxidoreductase subunit k;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 5 |
|
PDB 1a6q chain A domain 1
Region: 374 - 409
Aligned: 35
Modelled: 36
Confidence: 27.5%
Identity: 17%
Fold: Another 3-helical bundle
Superfamily: Protein serine/threonine phosphatase 2C, C-terminal domain
Family: Protein serine/threonine phosphatase 2C, C-terminal domain
Phyre2
| 6 |
|
PDB 2l3i chain A
Region: 298 - 309
Aligned: 12
Modelled: 12
Confidence: 18.5%
Identity: 50%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:aoxki4a, antimicrobial peptide in spider venom;
PDBTitle: oxki4a, spider derived antimicrobial peptide
Phyre2
| 7 |
|
PDB 1o8b chain B domain 1
Region: 301 - 337
Aligned: 34
Modelled: 37
Confidence: 17.9%
Identity: 15%
Fold: NagB/RpiA/CoA transferase-like
Superfamily: NagB/RpiA/CoA transferase-like
Family: D-ribose-5-phosphate isomerase (RpiA), catalytic domain
Phyre2
| 8 |
|
PDB 1s1q chain A
Region: 233 - 248
Aligned: 16
Modelled: 16
Confidence: 11.8%
Identity: 31%
Fold: UBC-like
Superfamily: UBC-like
Family: UEV domain
Phyre2
| 9 |
|
PDB 1bcc chain E domain 2
Region: 108 - 141
Aligned: 34
Modelled: 34
Confidence: 6.8%
Identity: 18%
Fold: Single transmembrane helix
Superfamily: ISP transmembrane anchor
Family: ISP transmembrane anchor
Phyre2
| 10 |
|
PDB 3d37 chain A domain 2
Region: 237 - 244
Aligned: 8
Modelled: 8
Confidence: 6.1%
Identity: 38%
Fold: Phage tail proteins
Superfamily: Phage tail proteins
Family: Baseplate protein-like
Phyre2