| 1 |
|
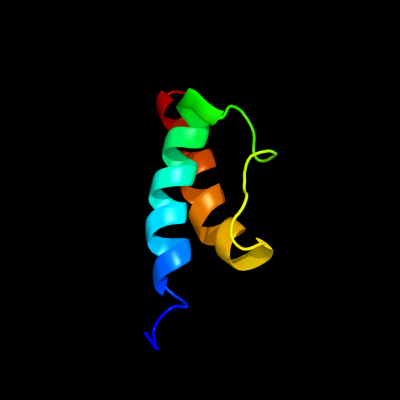
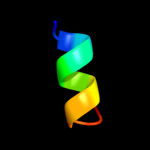
PDB 2jrm chain A
Region: 1 - 52
Aligned: 52
Modelled: 52
Confidence: 100.0%
Identity: 63%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ribosome modulation factor;
PDBTitle: solution nmr structure of ribosome modulation factor vp1593 from2 vibrio parahaemolyticus. northeast structural genomics target vpr55
Phyre2
| 2 |
|
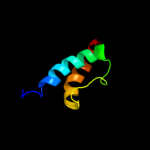
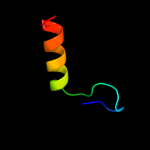
PDB 2rus chain B
Region: 11 - 49
Aligned: 39
Modelled: 39
Confidence: 13.0%
Identity: 21%
PDB header:lyase(carbon-carbon)
Chain: B: PDB Molecule:rubisco (ribulose-1,5-bisphosphate
PDBTitle: crystal structure of the ternary complex of ribulose-1,5-2 bisphosphate carboxylase, mg(ii), and activator co2 at 2.3-3 angstroms resolution
Phyre2
| 3 |
|
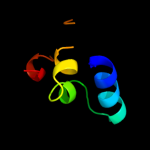
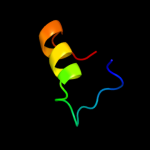
PDB 9rub chain B
Region: 11 - 49
Aligned: 39
Modelled: 39
Confidence: 13.0%
Identity: 23%
PDB header:lyase(carbon-carbon)
Chain: B: PDB Molecule:ribulose-1,5-bisphosphate carboxylase;
PDBTitle: crystal structure of activated ribulose-1,5-bisphosphate2 carboxylase complexed with its substrate, ribulose-1,5-3 bisphosphate
Phyre2
| 4 |
|
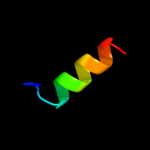
PDB 3bux chain B domain 3
Region: 32 - 47
Aligned: 16
Modelled: 16
Confidence: 9.9%
Identity: 19%
Fold: SH2-like
Superfamily: SH2 domain
Family: SH2 domain
Phyre2
| 5 |
|
PDB 2caz chain B
Region: 7 - 17
Aligned: 11
Modelled: 11
Confidence: 8.6%
Identity: 27%
PDB header:protein transport
Chain: B: PDB Molecule:vacuolar protein sorting-associated protein
PDBTitle: escrt-i core
Phyre2
| 6 |
|
PDB 2caz chain B domain 1
Region: 7 - 17
Aligned: 11
Modelled: 11
Confidence: 8.6%
Identity: 27%
Fold: Long alpha-hairpin
Superfamily: Endosomal sorting complex assembly domain
Family: VPS28 N-terminal domain
Phyre2
| 7 |
|
PDB 5rub chain A domain 1
Region: 11 - 49
Aligned: 39
Modelled: 39
Confidence: 8.6%
Identity: 23%
Fold: TIM beta/alpha-barrel
Superfamily: RuBisCo, C-terminal domain
Family: RuBisCo, large subunit, C-terminal domain
Phyre2
| 8 |
|
PDB 2wyb chain A
Region: 7 - 22
Aligned: 16
Modelled: 16
Confidence: 7.7%
Identity: 31%
PDB header:hydrolase
Chain: A: PDB Molecule:acyl-homoserine lactone acylase pvdq subunit
PDBTitle: the quorum quenching n-acyl homoserine lactone acylase pvdq2 with a covalently bound dodecanoic acid
Phyre2
| 9 |
|
PDB 2f6m chain B domain 1
Region: 7 - 17
Aligned: 11
Modelled: 11
Confidence: 7.7%
Identity: 27%
Fold: Long alpha-hairpin
Superfamily: Endosomal sorting complex assembly domain
Family: VPS28 N-terminal domain
Phyre2
| 10 |
|
PDB 1k0m chain A domain 2
Region: 18 - 43
Aligned: 26
Modelled: 26
Confidence: 7.6%
Identity: 31%
Fold: Thioredoxin fold
Superfamily: Thioredoxin-like
Family: Glutathione S-transferase (GST), N-terminal domain
Phyre2
| 11 |
|
PDB 1t1j chain A
Region: 29 - 36
Aligned: 8
Modelled: 8
Confidence: 7.0%
Identity: 38%
Fold: Flavodoxin-like
Superfamily: N-(deoxy)ribosyltransferase-like
Family: Hypothetical protein PA1492
Phyre2
| 12 |
|
PDB 3bit chain A
Region: 21 - 46
Aligned: 26
Modelled: 26
Confidence: 6.6%
Identity: 27%
PDB header:transcription
Chain: A: PDB Molecule:fact complex subunit spt16;
PDBTitle: crystal structure of yeast spt16 n-terminal domain
Phyre2
| 13 |
|
PDB 2ae3 chain A
Region: 7 - 22
Aligned: 16
Modelled: 16
Confidence: 5.9%
Identity: 31%
PDB header:hydrolase
Chain: A: PDB Molecule:glutaryl 7-aminocephalosporanic acid acylase;
PDBTitle: glutaryl 7-aminocephalosporanic acid acylase: mutational study of2 activation mechanism
Phyre2