1 c3sokB_
99.5
11
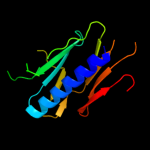
PDB header: cell adhesionChain: B: PDB Molecule: fimbrial protein;PDBTitle: dichelobacter nodosus pilin fima
2 d1oqwa_
99.5
15
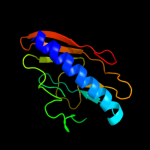
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pilin3 d2pila_
99.3
22
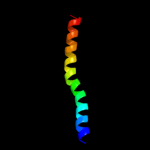
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pilin4 c2qv8B_
98.3
18
PDB header: transport proteinChain: B: PDB Molecule: general secretion pathway protein h;PDBTitle: structure of the minor pseudopilin epsh from the type 2 secretion2 system of vibrio cholerae
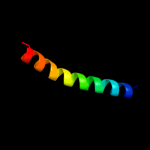
5 c2knqA_
97.3
14
PDB header: protein transportChain: A: PDB Molecule: general secretion pathway protein h;PDBTitle: solution structure of e.coli gsph
6 c4a18U_
32.3
55
PDB header: ribosomeChain: U: PDB Molecule: rpl13;PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of molecule 1
7 c3u5eL_
31.3
55
PDB header: ribosomeChain: L: PDB Molecule: 60s ribosomal protein l13-a;PDBTitle: the structure of the eukaryotic ribosome at 3.0 resolution
8 c2kncB_
16.3
5
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
9 d3ehbb2
13.1
26
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region10 c2kxeA_
12.7
43
PDB header: transferaseChain: A: PDB Molecule: dna polymerase ii small subunit;PDBTitle: n-terminal domain of the dp1 subunit of an archaeal d-family dna2 polymerase
11 d1fftb2
12.4
9
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region12 c1ybxA_
12.4
9
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: conserved hypothetical protein cth-383 from clostridium thermocellum
13 c2vnvC_
11.6
16
PDB header: sugar-binding proteinChain: C: PDB Molecule: bcla;PDBTitle: crystal structure of bcla lectin from burkholderia2 cenocepacia in complex with alpha-methyl-mannoside at 1.73 angstrom resolution
14 d1m56d_
10.4
25
Fold: Single transmembrane helixSuperfamily: Bacterial aa3 type cytochrome c oxidase subunit IVFamily: Bacterial aa3 type cytochrome c oxidase subunit IV15 d1puga_
10.0
9
Fold: YbaB-likeSuperfamily: YbaB-likeFamily: YbaB-like16 d1j8ba_
9.8
9
Fold: YbaB-likeSuperfamily: YbaB-likeFamily: YbaB-like17 c1qleB_
9.5
22
PDB header: oxidoreductase/immune systemChain: B: PDB Molecule: cytochrome c oxidase polypeptide ii;PDBTitle: cryo-structure of the paracoccus denitrificans four-subunit2 cytochrome c oxidase in the completely oxidized state3 complexed with an antibody fv fragment
18 c1ar1B_
9.5
22
PDB header: complex (oxidoreductase/antibody)Chain: B: PDB Molecule: cytochrome c oxidase;PDBTitle: structure at 2.7 angstrom resolution of the paracoccus2 denitrificans two-subunit cytochrome c oxidase complexed3 with an antibody fv fragment
19 c2kb1A_
9.0
9
PDB header: membrane proteinChain: A: PDB Molecule: wsk3;PDBTitle: nmr studies of a channel protein without membrane:2 structure and dynamics of water-solubilized kcsa
20 c2ljcA_
8.2
27
PDB header: transport protein/inhibitorChain: A: PDB Molecule: m2 protein, bm2 protein chimera;PDBTitle: structure of the influenza am2-bm2 chimeric channel bound to2 rimantadine
21 c2kadC_
not modelled
7.9
27
PDB header: membrane proteinChain: C: PDB Molecule: transmembrane peptide of matrix protein 2;PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain
22 c2kadA_
not modelled
7.9
27
PDB header: membrane proteinChain: A: PDB Molecule: transmembrane peptide of matrix protein 2;PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain
23 c2kadD_
not modelled
7.9
27
PDB header: membrane proteinChain: D: PDB Molecule: transmembrane peptide of matrix protein 2;PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain
24 c2kadB_
not modelled
7.9
27
PDB header: membrane proteinChain: B: PDB Molecule: transmembrane peptide of matrix protein 2;PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain
25 c2boiA_
not modelled
7.6
24
PDB header: lectinChain: A: PDB Molecule: cv-iil lectin;PDBTitle: 1.1a structure of chromobacterium violaceum lectin cv2l in2 complex with alpha-methyl-fucoside
26 d3dtub2
not modelled
7.6
16
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region27 c1bttA_
not modelled
7.5
27
PDB header: transmembrane proteinChain: A: PDB Molecule: band 3 anion transport protein;PDBTitle: the solution structures of the first and second2 transmembrane-spanning segments of band 3
28 c1btsA_
not modelled
7.5
27
PDB header: transmembrane proteinChain: A: PDB Molecule: band 3 anion transport protein;PDBTitle: the solution structures of the first and second2 transmembrane-spanning segments of band 3
29 c2hg5D_
not modelled
7.0
11
PDB header: membrane proteinChain: D: PDB Molecule: kcsa channel;PDBTitle: cs+ complex of a k channel with an amide to ester substitution in the2 selectivity filter
30 d1uzva_
not modelled
6.5
24
Fold: Calcium-mediated lectinSuperfamily: Calcium-mediated lectinFamily: Calcium-mediated lectin31 d1v54b2
not modelled
6.1
14
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region32 c2o01G_
not modelled
5.8
25
PDB header: photosynthesisChain: G: PDB Molecule: photosystem i reaction center subunit v,PDBTitle: the structure of a plant photosystem i supercomplex at 3.42 angstrom resolution
33 d1r3jc_
not modelled
5.8
12
Fold: Voltage-gated potassium channelsSuperfamily: Voltage-gated potassium channelsFamily: Voltage-gated potassium channels