| 1 |
|
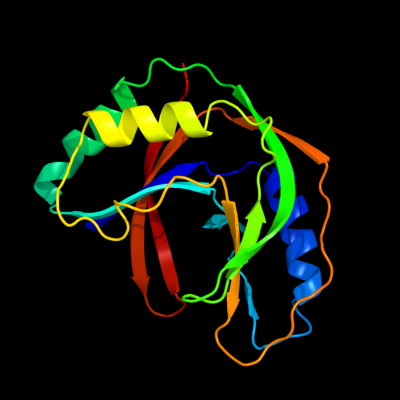
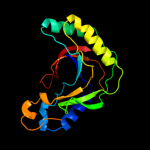
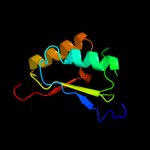
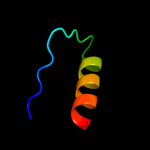
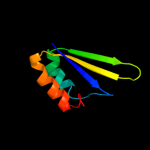
PDB 1iuh chain A
Region: 5 - 175
Aligned: 169
Modelled: 170
Confidence: 100.0%
Identity: 31%
Fold: LigT-like
Superfamily: LigT-like
Family: 2'-5' RNA ligase LigT
Phyre2
| 2 |
|
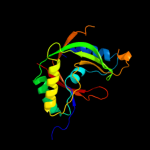
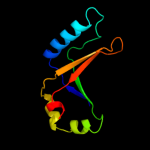
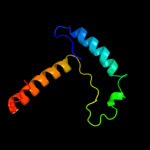
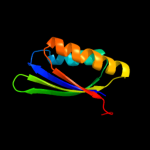
PDB 1vdx chain A
Region: 5 - 176
Aligned: 170
Modelled: 172
Confidence: 100.0%
Identity: 25%
PDB header:ligase
Chain: A: PDB Molecule:hypothetical protein ph0099;
PDBTitle: crystal structure of a pyrococcus horikoshii protein with2 similarities to 2'5' rna-ligase
Phyre2
| 3 |
|
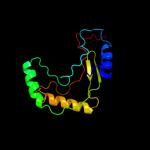
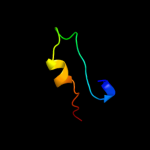
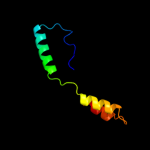
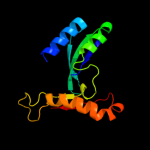
PDB 2d4g chain A
Region: 6 - 174
Aligned: 157
Modelled: 166
Confidence: 100.0%
Identity: 18%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein bsu11850;
PDBTitle: structure of yjcg protein, a putative 2'-5' rna ligase from2 bacillus subtilis
Phyre2
| 4 |
|
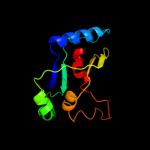
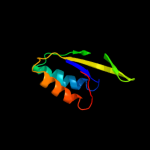
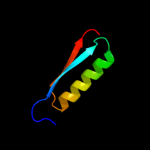
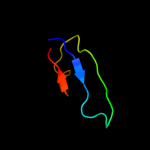
PDB 2vfy chain A
Region: 7 - 176
Aligned: 168
Modelled: 170
Confidence: 100.0%
Identity: 13%
PDB header:hydrolase
Chain: A: PDB Molecule:akap18 delta;
PDBTitle: akap18 delta central domain
Phyre2
| 5 |
|
PDB 1jh6 chain A
Region: 1 - 175
Aligned: 166
Modelled: 174
Confidence: 100.0%
Identity: 14%
Fold: LigT-like
Superfamily: LigT-like
Family: tRNA splicing product Appr>p cyclic nucleotide phosphodiesterase
Phyre2
| 6 |
|
PDB 2fsq chain A domain 1
Region: 11 - 139
Aligned: 125
Modelled: 129
Confidence: 98.2%
Identity: 12%
Fold: LigT-like
Superfamily: LigT-like
Family: Atu0111-like
Phyre2
| 7 |
|
PDB 2ilx chain A domain 1
Region: 7 - 129
Aligned: 118
Modelled: 123
Confidence: 87.0%
Identity: 18%
Fold: LigT-like
Superfamily: LigT-like
Family: 2',3'-cyclic nucleotide 3'-phosphodiesterase, catalytic domain
Phyre2
| 8 |
|
PDB 2e5c chain A
Region: 1 - 82
Aligned: 78
Modelled: 82
Confidence: 38.6%
Identity: 8%
PDB header:transferase
Chain: A: PDB Molecule:nicotinamide phosphoribosyltransferase;
PDBTitle: crystal structure of human nmprtase complexed with 5'-phosphoribosyl-2 1'-pyrophosphate
Phyre2
| 9 |
|
PDB 2i3e chain A
Region: 7 - 100
Aligned: 92
Modelled: 94
Confidence: 28.4%
Identity: 18%
PDB header:hydrolase
Chain: A: PDB Molecule:g-rich;
PDBTitle: solution structure of catalytic domain of goldfish rich2 protein
Phyre2
| 10 |
|
PDB 3h0d chain B
Region: 147 - 176
Aligned: 28
Modelled: 30
Confidence: 18.8%
Identity: 11%
PDB header:transcription/dna
Chain: B: PDB Molecule:ctsr;
PDBTitle: crystal structure of ctsr in complex with a 26bp dna duplex
Phyre2
| 11 |
|
PDB 3ce8 chain A
Region: 44 - 117
Aligned: 71
Modelled: 74
Confidence: 18.6%
Identity: 17%
PDB header:unknown function
Chain: A: PDB Molecule:putative pii-like nitrogen regulatory protein;
PDBTitle: crystal structure of a duf3240 family protein (sbal_0098) from2 shewanella baltica os155 at 2.40 a resolution
Phyre2
| 12 |
|
PDB 1gyx chain A
Region: 42 - 67
Aligned: 26
Modelled: 26
Confidence: 14.4%
Identity: 19%
Fold: Tautomerase/MIF
Superfamily: Tautomerase/MIF
Family: 4-oxalocrotonate tautomerase-like
Phyre2
| 13 |
|
PDB 2qpq chain C
Region: 46 - 118
Aligned: 69
Modelled: 73
Confidence: 11.0%
Identity: 13%
PDB header:transport protein
Chain: C: PDB Molecule:protein bug27;
PDBTitle: structure of bug27 from bordetella pertussis
Phyre2
| 14 |
|
PDB 3itf chain A
Region: 2 - 66
Aligned: 65
Modelled: 62
Confidence: 9.2%
Identity: 14%
PDB header:signaling protein
Chain: A: PDB Molecule:periplasmic adaptor protein cpxp;
PDBTitle: structural basis for the inhibitory function of the cpxp adaptor2 protein
Phyre2
| 15 |
|
PDB 3noh chain A
Region: 81 - 124
Aligned: 44
Modelled: 44
Confidence: 9.2%
Identity: 11%
PDB header:peptide binding protein
Chain: A: PDB Molecule:putative peptide binding protein;
PDBTitle: crystal structure of a putative peptide binding protein (rumgna_00914)2 from ruminococcus gnavus atcc 29149 at 1.60 a resolution
Phyre2
| 16 |
|
PDB 1aop chain A domain 2
Region: 6 - 71
Aligned: 59
Modelled: 66
Confidence: 8.9%
Identity: 15%
Fold: Ferredoxin-like
Superfamily: Nitrite/Sulfite reductase N-terminal domain-like
Family: Duplicated SiR/NiR-like domains 1 and 3
Phyre2
| 17 |
|
PDB 1u8s chain A domain 1
Region: 43 - 125
Aligned: 80
Modelled: 80
Confidence: 8.5%
Identity: 11%
Fold: Ferredoxin-like
Superfamily: ACT-like
Family: Glycine cleavage system transcriptional repressor
Phyre2
| 18 |
|
PDB 1zj8 chain B
Region: 6 - 127
Aligned: 114
Modelled: 122
Confidence: 7.8%
Identity: 10%
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable ferredoxin-dependent nitrite reductase nira;
PDBTitle: structure of mycobacterium tuberculosis nira protein
Phyre2
| 19 |
|
PDB 2ih2 chain A domain 2
Region: 123 - 156
Aligned: 33
Modelled: 34
Confidence: 7.8%
Identity: 18%
Fold: DNA methylase specificity domain
Superfamily: DNA methylase specificity domain
Family: TaqI C-terminal domain-like
Phyre2
| 20 |
|
PDB 3dku chain B
Region: 122 - 129
Aligned: 8
Modelled: 8
Confidence: 7.8%
Identity: 50%
PDB header:hydrolase
Chain: B: PDB Molecule:putative phosphohydrolase;
PDBTitle: crystal structure of nudix hydrolase orf153, ymfb, from2 escherichia coli k-1
Phyre2
| 21 |
|