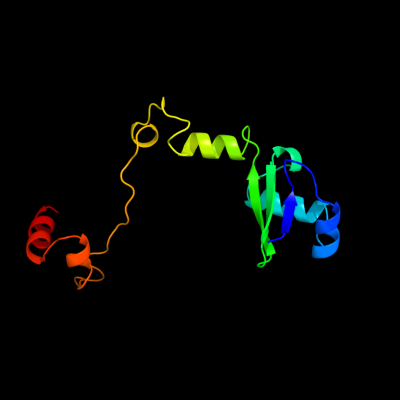
1 c3c6fD_
100.0
23
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: yetf protein;PDBTitle: crystal structure of protein bsu07140 from bacillus subtilis
2 c1yuzB_
53.2
21
PDB header: oxidoreductaseChain: B: PDB Molecule: nigerythrin;PDBTitle: partially reduced state of nigerythrin
3 d2guka1
43.5
15
Fold: PG1857-likeSuperfamily: PG1857-likeFamily: PG1857-like4 c2hr5B_
39.3
18
PDB header: metal binding proteinChain: B: PDB Molecule: rubrerythrin;PDBTitle: pf1283- rubrerythrin from pyrococcus furiosus iron bound form
5 d2e74d2
37.8
26
Fold: Single transmembrane helixSuperfamily: ISP transmembrane anchorFamily: ISP transmembrane anchor6 c2znmA_
33.9
21
PDB header: oxidoreductaseChain: A: PDB Molecule: thiol:disulfide interchange protein dsba;PDBTitle: oxidoreductase nmdsba3 from neisseria meningitidis
7 c3dvwA_
33.2
19
PDB header: oxidoreductaseChain: A: PDB Molecule: thiol:disulfide interchange protein dsba;PDBTitle: crystal structure of reduced dsba1 from neisseria2 meningitidis
8 c3gv1A_
30.2
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: disulfide interchange protein;PDBTitle: crystal structure of disulfide interchange protein from neisseria2 gonorrhoeae
9 c1dvbA_
27.0
26
PDB header: electron transportChain: A: PDB Molecule: rubrerythrin;PDBTitle: rubrerythrin
10 d1fvka_
25.5
15
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: DsbA-like11 c2kncA_
25.3
15
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
12 c3l9vE_
24.9
16
PDB header: oxidoreductaseChain: E: PDB Molecule: putative thiol-disulfide isomerase or thioredoxin;PDBTitle: crystal structure of salmonella enterica serovar typhimurium srga
13 d1e42a2
20.7
19
Fold: Subdomain of clathrin and coatomer appendage domainSuperfamily: Subdomain of clathrin and coatomer appendage domainFamily: Clathrin adaptor appendage, alpha and beta chain-specific domain14 d1z6ma1
19.5
21
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: DsbA-like15 d1un2a_
18.5
16
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: DsbA-like16 d2fwha1
18.2
22
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Thioltransferase17 d1eeja1
18.1
23
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: DsbC/DsbG C-terminal domain-like18 c2k1aA_
17.9
15
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
19 d1y5ib1
17.0
18
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins20 c2r77A_
15.9
14
PDB header: lipid binding proteinChain: A: PDB Molecule: phosphatidylethanolamine-binding protein, putative;PDBTitle: crystal structure of phosphatidylethanolamine-binding protein,2 pfl0955c, from plasmodium falciparum
21 c3kzqE_
not modelled
15.5
6
PDB header: structural genomics, unknown functionChain: E: PDB Molecule: putative uncharacterized protein vp2116;PDBTitle: the crystal structure of the protein with unknown function from vibrio2 parahaemolyticus rimd 2210633
22 c3feuA_
not modelled
15.4
13
PDB header: oxidoreductaseChain: A: PDB Molecule: putative lipoprotein;PDBTitle: crystal structure of dsba-like thioredoxin domain vf_a0457 from vibrio2 fischeri
23 c1t3bA_
not modelled
15.0
19
PDB header: isomeraseChain: A: PDB Molecule: thiol:disulfide interchange protein dsbc;PDBTitle: x-ray structure of dsbc from haemophilus influenzae
24 d2ewha1
not modelled
14.5
23
Fold: Ferredoxin-likeSuperfamily: CcmK-likeFamily: CcmK-like25 c3hd5A_
not modelled
13.7
16
PDB header: oxidoreductaseChain: A: PDB Molecule: thiol:disulfide interchange protein dsba;PDBTitle: crystal structure of a thiol:disulfide interchange protein2 dsba from bordetella parapertussis
26 c1jzdA_
not modelled
13.6
23
PDB header: oxidoreductaseChain: A: PDB Molecule: thiol:disulfide interchange protein dsbc;PDBTitle: dsbc-dsbdalpha complex
27 d1t3ba1
not modelled
13.5
19
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: DsbC/DsbG C-terminal domain-like28 d1iwga5
not modelled
12.6
12
Fold: Multidrug efflux transporter AcrB TolC docking domain; DN and DC subdomainsSuperfamily: Multidrug efflux transporter AcrB TolC docking domain; DN and DC subdomainsFamily: Multidrug efflux transporter AcrB TolC docking domain; DN and DC subdomains29 d1d4oa_
not modelled
11.8
19
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Transhydrogenase domain III (dIII)30 c1pt9B_
not modelled
11.8
19
PDB header: oxidoreductaseChain: B: PDB Molecule: nad(p) transhydrogenase, mitochondrial;PDBTitle: crystal structure analysis of the diii component of transhydrogenase2 with a thio-nicotinamide nucleotide analogue
31 c3gmfA_
not modelled
11.7
21
PDB header: oxidoreductaseChain: A: PDB Molecule: protein-disulfide isomerase;PDBTitle: crystal structure of protein-disulfide isomerase from novosphingobium2 aromaticivorans
32 d1pnoa_
not modelled
11.5
26
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Transhydrogenase domain III (dIII)33 d1aipc1
not modelled
11.2
16
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: TS-N domain34 d1dl6a_
not modelled
10.6
13
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain35 c3h93A_
not modelled
10.4
21
PDB header: transcription regulatorChain: A: PDB Molecule: thiol:disulfide interchange protein dsba;PDBTitle: crystal structure of pseudomonas aeruginosa dsba
36 d1beda_
not modelled
10.4
22
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: DsbA-like37 c2eelA_
not modelled
10.4
14
PDB header: apoptosisChain: A: PDB Molecule: cell death activator cide-a;PDBTitle: solution structure of the cide-n domain of human cell death2 activator cide-a
38 c2bruC_
not modelled
10.0
30
PDB header: oxidoreductaseChain: C: PDB Molecule: nad(p) transhydrogenase subunit beta;PDBTitle: complex of the domain i and domain iii of escherichia coli2 transhydrogenase
39 c3gykC_
not modelled
9.8
10
PDB header: oxidoreductaseChain: C: PDB Molecule: 27kda outer membrane protein;PDBTitle: the crystal structure of a thioredoxin-like oxidoreductase from2 silicibacter pomeroyi dss-3
40 d1ppva_
not modelled
9.6
12
Fold: NudixSuperfamily: NudixFamily: IPP isomerase-like41 d2cp9a1
not modelled
9.2
21
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: TS-N domain42 d1pfta_
not modelled
9.2
14
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain43 d1lkoa2
not modelled
9.0
30
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin44 d1ckma1
not modelled
8.7
26
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: DNA ligase/mRNA capping enzyme postcatalytic domain45 c3fz5C_
not modelled
8.6
23
PDB header: isomeraseChain: C: PDB Molecule: possible 2-hydroxychromene-2-carboxylate isomerase;PDBTitle: crystal structure of possible 2-hydroxychromene-2-carboxylate2 isomerase from rhodobacter sphaeroides
46 d1thta_
not modelled
8.4
13
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Thioesterases47 d1ibxa_
not modelled
8.4
25
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: CAD domain48 d1xb2b1
not modelled
8.3
21
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: TS-N domain49 d1yuza2
not modelled
8.0
32
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin50 d2fwua1
not modelled
7.9
18
Fold: Immunoglobulin-like beta-sandwichSuperfamily: CalX-likeFamily: CalX-beta domain51 c2d7cD_
not modelled
7.8
8
PDB header: protein transportChain: D: PDB Molecule: rab11 family-interacting protein 3;PDBTitle: crystal structure of human rab11 in complex with fip3 rab-2 binding domain
52 d1efub3
not modelled
7.7
32
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: TS-N domain53 c3gn3B_
not modelled
7.7
19
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative protein-disulfide isomerase;PDBTitle: crystal structure of a putative protein-disulfide isomerase from2 pseudomonas syringae to 2.5a resolution.
54 d1h6za3
not modelled
7.6
9
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: Pyruvate phosphate dikinase, N-terminal domain55 d2i5ia1
not modelled
7.5
16
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: YdjC-like56 d1t5la1
not modelled
7.5
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain57 d2b2na1
not modelled
7.4
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain58 c3kcmC_
not modelled
7.4
17
PDB header: oxidoreductaseChain: C: PDB Molecule: thioredoxin family protein;PDBTitle: the crystal structure of thioredoxin protein from geobacter2 metallireducens
59 d1d4ba_
not modelled
7.3
24
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: CAD domain60 d1nnqa2
not modelled
7.3
31
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin61 d1zd0a1
not modelled
7.2
18
Fold: PF0523-likeSuperfamily: PF0523-likeFamily: PF0523-like62 d2fi0a1
not modelled
7.2
8
Fold: SP0561-likeSuperfamily: SP0561-likeFamily: SP0561-like63 d2h7aa1
not modelled
7.2
17
Fold: YcgL-likeSuperfamily: YcgL-likeFamily: YcgL-like64 c3bciA_
not modelled
7.0
17
PDB header: oxidoreductaseChain: A: PDB Molecule: disulfide bond protein a;PDBTitle: crystal structure of staphylococcus aureus dsba
65 d2odka1
not modelled
6.8
25
Fold: YefM-likeSuperfamily: YefM-likeFamily: YefM-like66 d1meka_
not modelled
6.7
19
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: PDI-like67 d2gmga1
not modelled
6.6
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: PF0610-like68 c3rcqA_
not modelled
6.6
10
PDB header: oxidoreductaseChain: A: PDB Molecule: aspartyl/asparaginyl beta-hydroxylase;PDBTitle: crystal structure of human aspartate beta-hydroxylase isoform a
69 c2ivwA_
not modelled
6.5
18
PDB header: lipoproteinChain: A: PDB Molecule: pilp pilot protein;PDBTitle: the solution structure of a domain from the neisseria2 meningitidis pilp pilot protein.
70 c2odkD_
not modelled
6.4
25
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: hypothetical protein;PDBTitle: putative prevent-host-death protein from nitrosomonas europaea
71 c2x5rA_
not modelled
6.3
27
PDB header: viral proteinChain: A: PDB Molecule: hypothetical protein orf126;PDBTitle: crystal structure of the hypothetical protein orf126 from2 pyrobaculum spherical virus
72 c1ngmB_
not modelled
6.2
23
PDB header: transcription/dnaChain: B: PDB Molecule: transcription factor iiib brf1 subunit;PDBTitle: crystal structure of a yeast brf1-tbp-dna ternary complex
73 c2jugB_
not modelled
6.1
20
PDB header: biosynthetic proteinChain: B: PDB Molecule: tubc protein;PDBTitle: multienzyme docking in hybrid megasynthetases
74 d1szpb1
not modelled
6.1
29
Fold: SAM domain-likeSuperfamily: Rad51 N-terminal domain-likeFamily: DNA repair protein Rad51, N-terminal domain75 d1cy5a_
not modelled
6.0
5
Fold: DEATH domainSuperfamily: DEATH domainFamily: Caspase recruitment domain, CARD76 c2kvsA_
not modelled
5.6
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein mw0776;PDBTitle: nmr solution structure of q7a1e8 protein from staphylococcus2 aureus: northeast structural genomics consortium target:3 zr215
77 c1b22A_
not modelled
5.6
22
PDB header: dna binding proteinChain: A: PDB Molecule: dna repair protein rad51;PDBTitle: rad51 (n-terminal domain)
78 d1b22a_
not modelled
5.6
22
Fold: SAM domain-likeSuperfamily: Rad51 N-terminal domain-likeFamily: DNA repair protein Rad51, N-terminal domain79 c3c7mB_
not modelled
5.6
25
PDB header: oxidoreductaseChain: B: PDB Molecule: thiol:disulfide interchange protein dsba-like;PDBTitle: crystal structure of reduced dsbl
80 c1xtzA_
not modelled
5.5
10
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase;PDBTitle: crystal structure of the s. cerevisiae d-ribose-5-phosphate isomerase:2 comparison with the archeal and bacterial enzymes
81 c3ghaA_
not modelled
5.5
10
PDB header: oxidoreductaseChain: A: PDB Molecule: disulfide bond formation protein d;PDBTitle: crystal structure of etda-treated bdbd (reduced)
82 d1hyua4
not modelled
5.5
25
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: PDI-like83 c1e3hA_
not modelled
5.4
26
PDB header: polyribonucleotide transferaseChain: A: PDB Molecule: guanosine pentaphosphate synthetase;PDBTitle: semet derivative of streptomyces antibioticus pnpase/gpsi2 enzyme
84 c2jqeA_
not modelled
5.3
6
PDB header: signaling proteinChain: A: PDB Molecule: signal recognition 54 kda protein;PDBTitle: soution structure of af54 m-domain
85 c3hryA_
not modelled
5.3
35
PDB header: antitoxinChain: A: PDB Molecule: prevent host death protein;PDBTitle: crystal structure of phd in a trigonal space group and partially2 disordered
86 d1wpxb1
not modelled
5.1
13
Fold: PEBP-likeSuperfamily: PEBP-likeFamily: Phosphatidylethanolamine binding protein87 d1iwga6
not modelled
5.1
9
Fold: Multidrug efflux transporter AcrB TolC docking domain; DN and DC subdomainsSuperfamily: Multidrug efflux transporter AcrB TolC docking domain; DN and DC subdomainsFamily: Multidrug efflux transporter AcrB TolC docking domain; DN and DC subdomains88 c2oarA_
not modelled
5.1
12
PDB header: membrane proteinChain: A: PDB Molecule: large-conductance mechanosensitive channel;PDBTitle: mechanosensitive channel of large conductance (mscl)