| 1 |
|
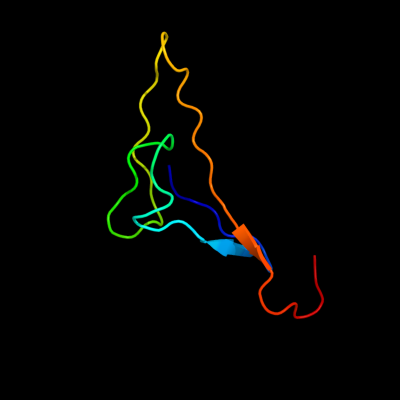
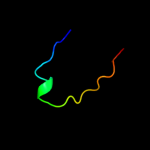
PDB 2kyw chain A
Region: 25 - 94
Aligned: 51
Modelled: 56
Confidence: 43.0%
Identity: 31%
PDB header:cell adhesion
Chain: A: PDB Molecule:adhesion exoprotein;
PDBTitle: solution nmr structure of a domain of adhesion exoprotein from2 pediococcus pentosaceus, northeast structural genomics consortium3 target ptr41o
Phyre2
| 2 |
|
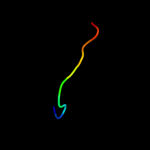
PDB 1zy3 chain B
Region: 73 - 83
Aligned: 11
Modelled: 11
Confidence: 28.4%
Identity: 64%
PDB header:apoptosis
Chain: B: PDB Molecule:bh3-peptide from bh3 interacting domain death
PDBTitle: structural model of complex of bcl-w protein with bid bh3-2 peptide
Phyre2
| 3 |
|
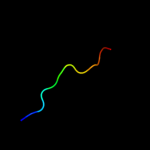
PDB 2oy7 chain A
Region: 24 - 55
Aligned: 32
Modelled: 32
Confidence: 16.8%
Identity: 38%
PDB header:membrane protein
Chain: A: PDB Molecule:outer surface protein a;
PDBTitle: the crystal structure of ospa mutant
Phyre2
| 4 |
|
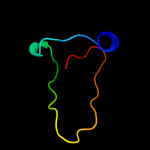
PDB 2voi chain B
Region: 70 - 83
Aligned: 14
Modelled: 14
Confidence: 16.0%
Identity: 50%
PDB header:apoptosis
Chain: B: PDB Molecule:bh3-interacting domain death agonist p13;
PDBTitle: structure of mouse a1 bound to the bid bh3-domain
Phyre2
| 5 |
|
PDB 1erf chain A
Region: 9 - 20
Aligned: 12
Modelled: 12
Confidence: 11.5%
Identity: 58%
PDB header:viral protein
Chain: A: PDB Molecule:transmembrane glycoprotein;
PDBTitle: conformational mapping of the n-terminal fusion peptide of2 hiv-1 gp41 using 13c-enhanced fourier transform infrared3 spectroscopy (ftir)
Phyre2
| 6 |
|
PDB 2pjv chain A
Region: 9 - 20
Aligned: 12
Modelled: 12
Confidence: 11.2%
Identity: 58%
PDB header:viral protein
Chain: A: PDB Molecule:envelope glycoprotein;
PDBTitle: solution structure of hiv-1 gp41 fusion domain bound to dpc2 micelle
Phyre2
| 7 |
|
PDB 3kd4 chain A
Region: 24 - 44
Aligned: 21
Modelled: 21
Confidence: 10.6%
Identity: 19%
PDB header:hydrolase
Chain: A: PDB Molecule:putative protease;
PDBTitle: crystal structure of a putative protease (bdi_1141) from2 parabacteroides distasonis atcc 8503 at 2.00 a resolution
Phyre2
| 8 |
|
PDB 1kwg chain A domain 3
Region: 31 - 40
Aligned: 10
Modelled: 10
Confidence: 8.2%
Identity: 50%
Fold: Flavodoxin-like
Superfamily: Class I glutamine amidotransferase-like
Family: A4 beta-galactosidase middle domain
Phyre2
| 9 |
|
PDB 2jpw chain A
Region: 61 - 72
Aligned: 12
Modelled: 12
Confidence: 6.7%
Identity: 58%
PDB header:contractile protein
Chain: A: PDB Molecule:troponin i, cardiac muscle;
PDBTitle: solution structure of the bisphosphorylated cardiac2 specific n-extension of cardiac troponin i
Phyre2
| 10 |
|
PDB 1jzt chain A
Region: 25 - 33
Aligned: 9
Modelled: 9
Confidence: 6.1%
Identity: 67%
Fold: YjeF N-terminal domain-like
Superfamily: YjeF N-terminal domain-like
Family: YjeF N-terminal domain-like
Phyre2
| 11 |
|
PDB 2wto chain B
Region: 11 - 44
Aligned: 34
Modelled: 34
Confidence: 6.1%
Identity: 29%
PDB header:metal binding protein
Chain: B: PDB Molecule:orf131 protein;
PDBTitle: crystal structure of apo-form czce from c. metallidurans ch34
Phyre2
| 12 |
|
PDB 2zkr chain 9
Region: 30 - 38
Aligned: 9
Modelled: 9
Confidence: 6.0%
Identity: 44%
PDB header:ribosomal protein/rna
Chain: 9: PDB Molecule:60s ribosomal protein l32;
PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
Phyre2
| 13 |
|
PDB 2ari chain A
Region: 9 - 20
Aligned: 12
Modelled: 12
Confidence: 5.6%
Identity: 58%
PDB header:viral protein
Chain: A: PDB Molecule:envelope polyprotein gp160;
PDBTitle: solution structure of micelle-bound fusion domain of hiv-12 gp41
Phyre2